✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
QuantiTect Virus Kit (1000)
Cat. No. / ID: 211015
✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
特点
- 高灵敏度的单重和多重检测
- 在同一反应中检测病毒 RNA 或 DNA
- 明确地检测微弱的阳性信号
- 快速的通用 2 步法实验方案
- 5x 浓度预混液,用于通过提高样品的投入量而提高灵敏度
产品详情
绩效
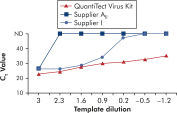
使用 QuantiTect Virus Kit 进行扩增时,即使模板量低而 CT 值高,也能在一定稀释度范围内呈现陡峭的 S 形曲线(请参阅图“ 在宽动态范围内明确测定 CT 值”)。这样就能在 Real-time PCR 中准确测定病毒核酸定量的 CT 值。
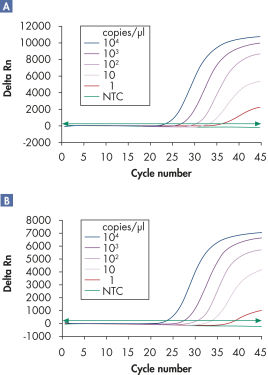
多重检测可在宽线性范围内检测多个病毒 RNA 或 DNA 靶标及内质控而不会降低灵敏度(请参阅图“ 在宽线性范围内可靠地检测病毒 RNA”和“ 改善了低载量病毒 RNA 的检测”)。
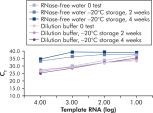
试剂盒附带的 QuantiTect Nucleic Acid Dilution Buffer 可在稀释和反应设置过程中稳定 RNA 和 DNA 标准品,并防止反应管或移液器吸头等塑料表面的核酸残留。该缓冲液可对用于病毒核酸定量的标准品进行可靠的稀释,从而提供从低到高 CT 值的宽线性范围,并确保标准品长期保存而不会发生降解(请参阅图“ 可靠的 RNA 标准品稀释和存放”)。
查看图表
原理
QuantiTect Virus Kit 可在单重或多重检测中首次尝试便能实现高灵敏度的病毒核酸检测(请参阅流程图“ QIAGEN 多重试剂盒”)。经优化的预混液可确保多重反应中 PCR 产物的扩增效率和灵敏度与相应单重扩增反应中的 PCR 产物相同。
在同一而非独立反应中扩增对照品和靶标基因可尽量减少操作错误,从而提高基因定量的可靠性。QuantiTect Virus Buffer 包含 K+ 和 NH4+ 离子的均衡组合以及独特的 Synthetic Factor MP,它们能共同促进引物和探针稳定且高效地退火到核酸模板,从而提高 PCR 的效率(请参阅图“ 独特的 PCR 缓冲液”)。此外,Sensiscript Reverse Transcriptase 的独特配方可确保病毒 RNA 的高灵敏度逆转录,而 HotStarTaq Plus DNA Polymerase 可提供严格的热启动,防止形成非特异性产物。
| 试剂盒组成 | 特点 | 优势 | |
|---|---|---|---|
| 5x QuantiTect Virus Master Mix | 浓缩预混液 | 高度浓缩,专为灵敏的病毒检测而优化 | 可在检测中加入更大体积的模板以提高灵敏度 |
| HotStarTaq Plus DNA Polymerase | 在 95ºC 下 5 分钟活化 | 在室温下配制 qPCR 反应 | |
| QuantiTect Virus Buffer | NH4+ 和 K+ 离子的平衡组合 | 特异性引物退火确保可靠的 PCR 结果 | |
| Synthetic Factor MP | 可在一管反应中对多达 4 个基因进行可靠的多重分析 | ||
| 其他试剂盒组分 | QuantiTect Virus RT Mix | 含有 Sensiscript Reverse Transcriptase 的独特配方 | 针对病毒 RNA 的高灵敏度检测进行了优化 |
| QuantiTect 核酸稀释缓冲液 | 用于稀释和保存核酸标准品的专有缓冲液制剂。 | 在稀释和反应设置过程中稳定 RNA 和 DNA 标准品,并防止反应管或移液器吸头等塑料表面的核酸残留 |
查看图表
程序
QuantiTect Virus Kit 使用序列特异性探针对病毒核酸(RNA 和/或 DNA)和内质控进行高灵敏度的 Real-time PCR 分析。反应可在包含或不包含逆转录步骤的情况下进行,从而可灵活设计多重检测来检测 RNA 靶标、DNA 靶标或同时检测 RNA 和 DNA 靶标。请按照手册中的方案来获得快速可靠的结果。
试剂盒提供时预混液中可包含或不含 ROX 参比荧光染料(见表)。
| ROX 染料 | 试剂盒 | 兼容的定量 PCR 仪 |
|---|---|---|
| 在预混液中提供 | QuantiTect Virus Kit | Applied Biosystems 除 Applied Biosystems 7500 外的所有定量 PCR 仪 |
| 以单管形式提供 | QuantiTect Virus +ROX Vial Kit | Applied Biosystems 7500 和来自 Bio-Rad、Cepheid、Eppendorf、QIAGEN、Roche、Agilent 及其他供应商的定量 PCR 仪 |
对于快速、高灵敏度的终点一步法 RT-PCR 应用(包括病毒检测),我们建议使用 QIAGEN OneStep RT-PCR Kit。
应用
辅助数据和图表
在宽动态范围内明确测定 CT 值。
Specifications
| Features | Specifications |
|---|---|
| Applications | 病毒检测 |
| SYBR Green I or sequence-specific probes | 序列特异性探针 |
| Real-time or endpoint | 实时 |
| Reaction type | 逆转录和 PCR |
| Thermal cycler | 大多数实时定量 PCR 仪(毛细管定量 PCR 仪除外,例如 LightCycler® 1.x 和 2.0) |
| Sample/target type | RNA 或 DNA 靶标 |
| With or without ROX | 可以含 ROX 的预混液以及独立小管包装的 ROX 形式提供 |
| Single or multiplex | 单重或多重 |