特点
- QIAGEN PCR缓冲体系最大限度地减少了PCR条件的优化
- 预混液规格易于反应体系构建
- 更少的移液操作最大限度减少污染风险
产品详情
绩效
Taq DNA Polymerase规格
浓度:5单位/µl
重组酶:有
底物类似物:dNTP、ddNTP、dUTP、biotin-11-dUTP、DIG-11-dUTP、荧光-dNTP/ddNTP
延伸率:72°C下2–4 kb/分钟
半衰期:97°C下10分钟;94°C下60分钟
扩增效率:≥105倍
5'–>3'外切酶活性:有
Extra A辅助剂: 有
3'–>5'外切酶活性:无
污染核酸:无
污染RNA酶:无
污染蛋白酶:无
自吸泵活性:无
查看图表
原理
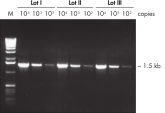
Taq PCR Master Mix Kit包含预混合规格的QIAGEN's Taq DNA Polymerase。这种易于使用的反应液同样包含QIAGEN PCR Buffer、MgCl2和具优化浓度的超纯dNTP。只需加入引物和模板DNA即可构建PCR反应体系。由于方便的预混液规格,最大限度地减少了移液失误,确保了高重复的PCR结果(参见" Reproducible PCR")。Taq PCR Master Mix可在2–8°C下储存长达2个月,通过去除解冻时间来构建更快速的PCR反应体系。
Taq DNA Polymerase
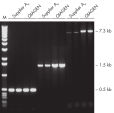
Taq DNA Polymerase是高品质的重组酶,适用于常规的和特殊的PCR应用(参见" Tolerance of different primer Tm values"和" Specific amplification of long PCR products")。
QIAGEN PCR Buffer
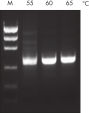
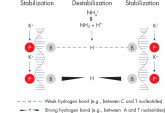
研发新型的QIAGEN PCR Buffer通过减少使用时对PCR条件的优化,来节省时间和精力。QIAGEN PCR Buffer包括KCl和(NH4)2SO4。独特的缓冲体系有助于特异性PCR产物的扩增。在每个PCR循环的退火步骤中,该缓冲液具有特异性-非特异性引物的高比率结合。由于KCl和(NH4)2SO4独特的平衡结合,相比传统的PCR缓冲液,该PCR缓冲液提供更大范围退火温度和Mg2+浓度条件,以及严格的引物退火条件。通过改变退火温度或Mg2+浓度,即可显著减少PCR优化过程,而且通常不需要优化(参见" Wide annealing temperature window"和" Tolerance to variable magnesium concentration")。
查看图表
程序
应用
Taq PCR Master Mix Kit适用于常规的和特殊的应用,包括:
- 常规的PCR
- RT-PCR
- 筛选
- 基于PCR的DNA指纹图谱分析(VNTR、STR和RAPD)
辅助数据和图表
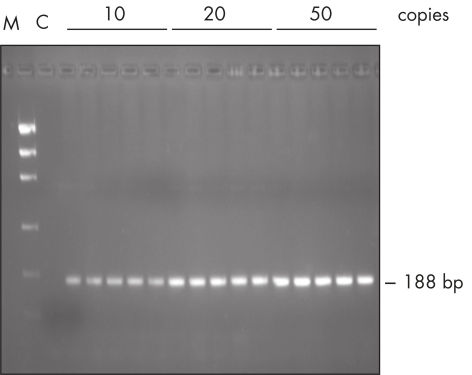
Reproducible PCR.
Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, RT-PCR, DNA fingerprinting |
| dNTP's included | Yes (in Master Mix) |
| Mastermix | Yes |
| Reaction type | PCR amplification |
| Enzyme activity | 5' -> 3' exonuclease activity |
| Real-time or endpoint | Endpoint |
| Sample/target type | Genomic DNA and cDNA |
| Single or multiplex | Single |
| With/without hotstart | Without hotstart |