Taq DNA Polymerase (250 U)
Cat. No. / ID: 201203
特点
- QIAGEN PCR缓冲体系最大限度地减少了PCR条件的优化
- 独特的即用型PCR缓冲液,操作更快速简单
- Q-Solution可帮助扩增GC含量高的模板
- 提供多种规格的包装方便使用
产品详情
绩效
Taq DNA Polymerase可在多种PCR条件下进行灵敏的PCR反应,无需耗时的优化过程(参见" Tolerance of different primer Tm Values" 和" Specific amplification of long PCR products")。每批Taq DNA Polymerase都受到全面的质量控制检测,包括严格的PCR特异性和可重复性分析,从人类基因组DNA扩增低拷贝的目的基因(参见" Lot-to-lot reproducibility")。试剂盒提供的QIAGEN PCR Buffer和CoralLoad PCR Buffer具有独特的组成,可在多种PCR条件下进行高度特异性的PCR,无需优化(参见" Wide annealing-temperature window" 和" Tolerance to variable magnesium concentration")。此外,CoralLoad PCR Buffer使得PCR产物可直接上样到琼脂糖凝胶,更易于操作,更快获得结果。试剂盒中提供的Q-Solution可进一步提高PCR的性能(参见" Amplification of difficult templates")。
Taq DNA Polymerase的规格
浓度: 5 单位/µl
重组酶: 是
底物类似物: dNTP、ddNTP、dUTP、biotin-11-dUTP、DIG-11-dUTP和fluorescent-dNTP/ddNTP
延伸速率: 72°C 2–4 kb/min
半衰期: 97°C 10 min;94°C 60 min
扩增效率: ≥105倍
5'–>3'外切酶活性: 有
额外添加A: 有
3'–>5'外切酶活性: 无
核酶污染: 无
RNases污染: 无
蛋白酶污染: 无
自引发活性: 无
查看图表
 Specific amplification of long PCR products.
Specific amplification of long PCR products.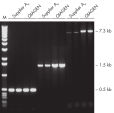 Lot-to-lot reproducibility.
Lot-to-lot reproducibility.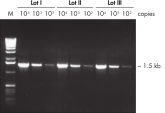 NH4+ and K+ cations in QIAGEN PCR buffers increase specific primer annealing.
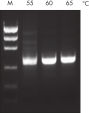
NH4+ and K+ cations in QIAGEN PCR buffers increase specific primer annealing.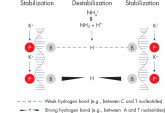 A. Wide annealing temperature window. B. Tolerance of variable magnesium concentration.
A. Wide annealing temperature window. B. Tolerance of variable magnesium concentration.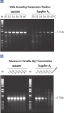 Amplification of difficult templates with Q-Solution.
Amplification of difficult templates with Q-Solution.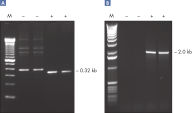
原理
Taq DNA Polymerase是一种高品质重组酶,适合常规和专门的PCR应用(参见" Tolerance of different primer Tm Values" 和" Specific amplification of long PCR products")。
QIAGEN PCR Buffer
研发的新型QIAGEN PCR Buffer可节省时间和精力,减少所需PCR优化。QIAGEN PCR Buffer含有KCl和(NH4)2SO4 。独特的缓冲液便于扩增特异性PCR产物。在每个PCR循环的退火步骤,缓冲液使引物结合具有高特异性比率。KCl和(NH4)2SO4独特比例结合,使PCR缓冲液与常规PCR缓冲液相比,可在更宽范围的退火温度和Mg2+浓度下提供严格的引物退火条件。极大减少通过改变退火温度或Mg2+浓度的PCR优化过程,有时甚至不需要(参见" Wide annealing temperature window" 和" Tolerance to variable magnesium concentration")。
CoralLoad PCR Buffer
CoralLoad PCR Buffer具有QIAGEN PCR Buffer的所有优点。此外,还可直接将PCR反应液上样到琼脂糖凝胶,无需再单独添加凝胶上样缓冲液。与常规QIAGEN PCR Buffer一样,CoralLoad PCR Buffer具有相同的PCR特异性和最少的反应优化。另外,缓冲液还含有两种标记染料:一种橙色染料和一种红色染料,便于估计DNA迁移距离和优化琼脂糖凝胶电泳时间(参见" CoralLoad PCR Buffer")。缓冲液提高了移液可视性,使PCR产物可直接上样到凝胶,提高了便利性。
Q-Solution
Q-Solution通过修饰DNA的熔解行为,便于扩增GC含量高的模板或含有高度二级结构的模板。使用这种独特的试剂常常能完成或改进不理想的PCR(参见" Amplification of difficult templates")。与DMSO和其他PCR添加剂不同,Q-Solution可在多种引物-模板体系中使用特定工作浓度,而不会产生毒性作用。
查看图表
 Specific amplification of long PCR products.
Specific amplification of long PCR products. NH4+ and K+ cations in QIAGEN PCR buffers increase specific primer annealing.
NH4+ and K+ cations in QIAGEN PCR buffers increase specific primer annealing. A. Wide annealing temperature window. B. Tolerance of variable magnesium concentration.
A. Wide annealing temperature window. B. Tolerance of variable magnesium concentration. CoralLoad PCR Buffer.
CoralLoad PCR Buffer.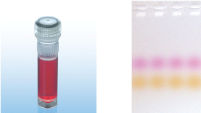 Amplification of difficult templates with Q-Solution.
Amplification of difficult templates with Q-Solution.
程序
应用
Taq DNA Polymerase适用于常规和特殊的应用,包括:
- 常规PCR
- RT-PCR
- 筛选
- 基于PCR的DNA指纹图谱分析(VNTR、STR和RAPD)
辅助数据和图表
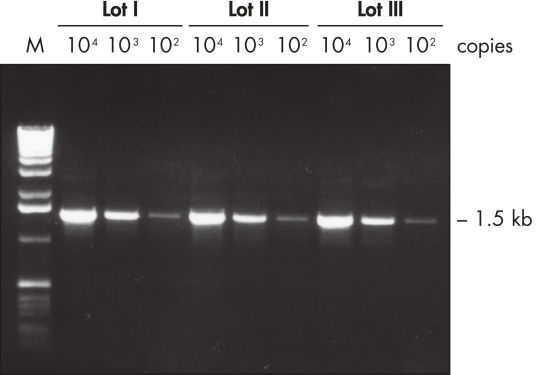
Lot-to-lot reproducibility.
A fragment of the single-copy gene for cystic fibrosis was amplified from 30 ng, 3 ng, and 300 pg human genomic DNA corresponding to 104, 103, and 102 copies of target template, respectively. Three different lots of QIAGEN Taq DNA Polymerase were used and equal volumes of the PCR product were analyzed on a 1% agarose gel. M: markers.
Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, RT-PCR, DNA fingerprinting |
| dNTP's included | No |
| Real-time or endpoint | Endpoint |
| Reaction type | PCR amplification |
| Single or multiplex | Single |
| With/without hotstart | Without hotstart |
| Enzyme activity | 5' -> 3' exonuclease activity |
| Mastermix | No |
| Sample/target type | Genomic DNA and cDNA |