✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
REPLI-g Single Cell Kit (24)
Cat. No. / ID: 150343
✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
特点
- 采用多重置换扩增技术对基因组位点进行无偏差的扩增
- 适用于二代测序等新技术应用
- 产量稳定,可达40 µg(产物平均长度大于>10 kb)
- 可用于癌症、干细胞或宏观基因组学(metagenomics)研究
产品详情
很多研究人员使用二代测序仪器对生物样本的DNA序列进行分析和基因分型,但经常受限于有限的样本量。REPLI-g Single Cell Kit可均一的扩增单个细胞(1至1000个细菌或肿瘤细胞)或纯化的基因组DNA,能够覆盖基因组所有位点。另有专用于干血或新鲜或冷冻的组织样本的实验方案。所有缓冲液和试剂生产都经过严格控制的流程,避免污染DNA,确保每次实验获得可靠的结果。该产品采用多重置换扩增(MDA)技术,对所有基因组位点进行无偏差的扩增。与基于PCR的全基因组扩增技术相比,该技术具有更好的扩增高保真度,避免出现假阳性或假阴性信号。
绩效
覆盖基因组所有位点,十分适用于二代测序和其他下游应用
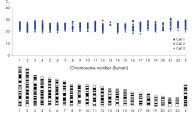
REPLI-g Single Cell Kit扩增的DNA平均长度大于10 kb,并且覆盖所有基因组位点。该产品已经过验证,适用于二代测序、基于芯片技术的比较基因组杂交(aCGH)和real-time PCR应用等多种下游分析。使用该产品无需单独进行PCR扩增,减少了手动操作,获得的产物长度PCR方法更长(参见" Next-generation sequencing using REPLI-g amplified DNA requires less hands-on time and generates more sequence information than PCR-based methods")。用扩增产物进行二代测序,获得高品质结果。这表明该试剂盒的位点覆盖率大,错配率低,其扩增的DNA(甚至从单一细菌细胞中扩增的DNA)能够与纯化获得的基因组DNA相媲美(参见" Comparable NGS results")。另有研究对人类常染色体和X染色体上的多个标记物进行了分析,三个独立的实验均显示:基因组的所有位点被成功扩增,没有遗漏任何一个位点(参见" Complete genome coverage")。
| 样本类型(细胞/DNA) | 研究领域 |
|---|---|
| 人类/动物 | 生物标记物研究(SNP、突变、CNV) |
| 干细胞研究 | |
| 循环胚胎细胞分析 | |
| 嵌合体研究 | |
| 遗传易感性研究 | |
| 转基因动物基因分型 | |
| 癌症 | 体细胞遗传变异分析 |
| 肿瘤恶化 | |
| 肿瘤干细胞/进化 | |
| 循环肿瘤细胞分析 | |
| 细菌 | 宏观基因组学研究 |
| 病原体分析 | |
| 微生物基因分型 | |
| Plants* | 植物气孔研究 |
| 花粉分析 |
REPLI-g Single Cell Kit的表现优于其他供应商的试剂盒
其他供应商的试剂盒基于PCR技术进行全基因组扩增,获得的产物片段较短,带有PCR引物序列,可能影响下游应用。基于PCR的全基因组扩增容易出现错误,会导致碱基对突变、STR的减少或增多,使用低保真Taq DNA聚合酶会导致位点缺失。相反,REPLI-g Single Cell Kit能够进行高度均一的全基因组扩增,位点偏差小。对四个试剂盒进行了检测,其中两个利用多重置换扩增技术(包括REPLI-g Single Cell Kit),另外两个利用PCR技术。采用单一细胞扩增实验方案对所有试剂盒进行序列位点检测。除REPLI-g Single Cell Kit外的其他供应商的试剂盒都出现了位点的遗漏(参见" Unbiased DNA amplification from a single cell")。
查看图表
原理
REPLI-g Single Cell Kit含有REPLI-g sc Polymerase,这是一种优化的新型高保真Phi 29聚合酶。该试剂盒采用多重置换扩增技术扩增复杂的基因组DNA,温和的碱孵育能够避免DNA片段化,促进对所有位点的无偏差扩增。该试剂盒十分适用于分离的肿瘤细胞或细菌细胞等单一细胞,能够扩增获得高产量DNA(参见表1)。此外,该试剂盒还配有专用于鲜血或干血、新鲜或冷冻组织的实验方案,可用于多种研究样本。常规DNA产量可达40 µg,对模板的起始量要求低,因此进行后续遗传分析时无需检测DNA浓度。REPLI-g Single Cell Kit的扩增产物平均长度超过10 kb,且覆盖所有位点,适合于多种应用,包括二代测序、基于芯片技术的比较基因组杂交、焦磷酸测序和real-time PCR分析(参见表2)。
| 应用 | 仪器 |
|---|---|
| 全基因组测序 | 二代测序平台† |
| 外显子组测序 | |
| SNP基因分型芯片 | 芯片平台† |
| Array CGH | |
| qPCR/PCR技术 | Real-time PCR/PCR仪† |
| Sanger测序 | 毛细管测序仪† |
| 焦磷酸测序 | PyroMark (QIAGEN) |
扩增原理
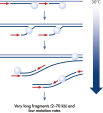
REPLI-g Single Cell Kit采用等温基因组扩增技术,即多重置换扩增(MDA)。六聚体与变性DNA随机结合,然后在等温条件下,在优化的Phi 29聚合酶作用下,发生链置换合成反应。每个置换后的单链作为模板,与引物结合,可扩增获得高产量DNA(参见" Multiple Displacement Amplification (MDA) technology")。Phi 29聚合酶为噬菌体衍生酶,具有3'→5'引物外切酶活性(校正活性),其保真性是Taq DNA聚合酶的1000倍。Phi 29聚合酶在优化的REPLI-g Single Cell缓冲液体系中,能够轻松的打开发卡结构等二级结构,因此可促进扩增时聚合酶正常工作。可扩增获得长达100 kb的DNA片段(参见" Unbiased amplification with Phi 29 polymerase")。
细胞裂解和DNA的碱变性
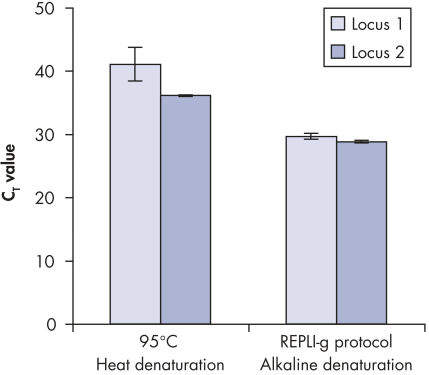
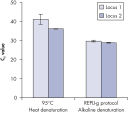
基因组DNA在用于酶促扩增反应前必须经过变性,而这一变性过程通常使用高温或高pH值(强碱)孵育。REPLI-g Single Cell Kit采用温和的碱孵育,在细胞裂解、DNA变性的同时,确保DNA片段化程度小,不产生碱基突变。因此扩增获得的DNA完整度高,产物长度长,覆盖位点多(参见" Effect of heat and alkaline denaturation on loci representation")。
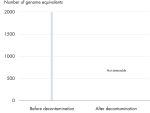
高效去除可检测到的DNA污染
REPLI-g Single Cell Kit中的所有组分都经过了独特的去污染流程,避免扩增产物被污染。在标准化流程中,对缓冲液和试剂进行紫外线照射,确保其不会污染DNA(参见" Innovative decontamination procedure")。紫外线照射后,对试剂盒的所有组分进行严格的质量控制,确保其性能。
查看图表
程序
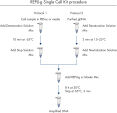
简单的单管操作
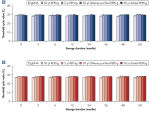
REPLI-g Single Cell Kit可对单一细胞或有限的样本进行简便、可靠、准确的全基因组扩增。反应体系构建十分便利、可靠,仅需15分钟左右的手动操作(参见" REPLI-g Single Cell Kit procedure")。专用的缓冲液和试剂可对单一细胞、有限的组织材料、或纯化的DNA进行扩增,产量高、位点覆盖率大(参见表3)。REPLI-g技术扩增获得的DNA可在–20°C长期储存(参见" Consistent long-term stability")。请参见表4,了解REPLI-g产品线的更多信息。
| 试剂盒成分 | 优势 |
|---|---|
| REPLI-g sc Polymerase | 最长可达70 kb的长片段 |
| 保真性是Taq聚合酶的1000倍 | |
| 覆盖所有位点 | |
| 均一扩增所有位点 | |
| REPLI-g sc Reaction Buffer‡ | 对所有位点进行无偏差的扩增 |
| Buffer DLB (裂解和变性) | 高效扩增 |
| 不损伤DNA | |
| 紫外线照射去除污染 | 减少可检测到的残留DNA污染 |
| REPLI-g Single Cell | REPLI-g Mini | REPLI-g UltraFast Mini | REPLI-g Midi | REPLI-g Screening | REPLI-g FFPE | REPLI-g Mitochondrial DNA | |
|---|---|---|---|---|---|---|---|
| 起始材料 | 单一细胞、gDNA | 纯化的基因组DNA、血液、细胞 | 纯化的基因组DNA、血液、细胞 | FFPE组织、FFPE组织中纯化的基因组DNA | 纯化的基因组DNA | ||
| 另外提供其他起始材料的实验方案 | |||||||
| 起始量 | 单一细胞、2–1000各细胞、组织、纯化的gDNA (1–10 ng) | >10 ng gDNA、0.5 µl血液或细胞(>600个细胞/µl) | >10 ng gDNA、0.5 µl血液或细胞(>600个细胞/µl) | 组织切片(直径1 cm,厚度10–40 µm);>100 ng gDNA | >1 ng纯化的gDNA | ||
| 产量(µg/反应) | 40 | 10 | 7–10 | 40 | 8 | 标准产量:≤10; 高产量:≤40 | 3–5 |
| 反应时间 | 8–16小时 | 10–16 小时 | 1.5小时 | 8–16小时 | 12–16小时 | 标准产量:4小时;高产量:10小时 | 8小时 |
| 手动操作时间 | 15分钟 | 15分钟 | 15分钟 | 15分钟 | 15分钟 | 40分钟 | 15分钟 |
| 规格 | 反应管 | 反应管 | 反应管 | 反应管 | 孔板 | ||
查看图表
应用
- 二代测序
- 采用TaqMan引物/探针对进行的SNP基因分型
- 基于qPCR和PCR的突变检测
- STR/微卫星分析
- Sanger测序
- 焦磷酸测序
- aCGH等芯片技术
辅助数据和图表
Effect of heat and alkaline denaturation on loci representation.
Effect of heat and alkaline denaturation on loci representation.