QIAseq Targeted RNA Indexes
用于对目标RNA测序所需样本和对QIAseq Targeted RNA Panels生成的RNA文库测序进行索引
QIAseq Targeted RNA Indexes
用于对目标RNA测序所需样本和对QIAseq Targeted RNA Panels生成的RNA文库测序进行索引

Cat. No. / ID: 333777

Cat. No. / ID: 333764

Cat. No. / ID: 333714

Cat. No. / ID: 333727

Cat. No. / ID: 333737

Cat. No. / ID: 333747

Cat. No. / ID: 333757

Cat. No. / ID: 333715

Cat. No. / ID: 333716

Cat. No. / ID: 333725

Cat. No. / ID: 333735
QIAseq Targeted Panels enable Sample to Insight next-generation sequencing (NGS) of DNA or RNA. These optimized solutions facilitate ultrasensitive variant and clonotype detection from cells, tissue and biofluids using integrated Unique Molecular Indices (UMIs).
In order to de-multiplex pooled libraries and assign reads to the appropriate original source library, NGS workflows require the addition of an adapter complex that includes both a common nucleotide sequence, as well as sample index oligonucleotides.
Compatible only with Illumina instruments, our Unique Dual Indexes (UDIs) are optimized for QIAseq Targeted Panel workflows, including QIAseq Targeted DNA, QIAseq RNAscan and QIAseq Immune Repertoire RNA Library Kits. Exclusively available from QIAGEN for QIAseq targeted NGS applications, QIAseq UDI Kits provide the highest level of process safety and ensure confidence in sequencing data by mitigating “index hopping” and mis-assignment of reads. The combination of both UMIs and UDIs provide QIAseq Targeted Panels with a remarkable level of accuracy. Combinatorial, non-unique dual index adapters are also available.
Need a quote for your research project or would you like to discuss your project with our specialist team? Contact Us
PCR duplicates are a major issue in targeted DNA sequencing, since, through PCR amplification, they turn unique DNA molecules into identical DNA molecules that cannot be distinguished from each other. In addition, errors from PCR amplification and sequencing process may also be present in final reads that lead to false-positive variants in sequencing results. This, in turn, results in the inability to confidently call DNA variants present at low frequencies in the starting DNA material. To overcome the issue of PCR duplicates and amplification artifacts, the QIAseq Targeted DNA Panels use digital sequencing by incorporating molecular barcodes into the starting DNA material before any amplification takes place, thereby preserving the uniqueness of the starting DNA molecules and overcoming the issues of PCR duplicates, false positives and library bias.
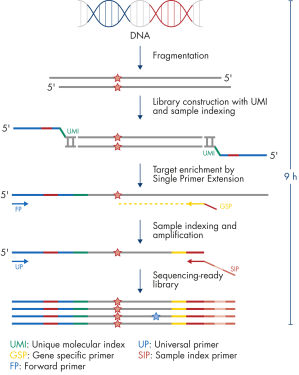
The entire workflow of the QIAseq Targeted DNA Panels to go from extracted DNA to sequencing-ready libraries can be completed in 9 hours (see figure Workflow). Extracted DNA is fragmented, genomic targets are molecularly barcoded and enriched, and libraries are constructed. Sequencing files can be fed into the QIAseq pipeline, a cloud-based data analysis pipeline, which will filter, map and align reads, as well as count unique molecular barcodes associated with targeted genomic regions, and call variants with a barcode-aware algorithm. This data can then be fed into IVA or QCI for interpretation.
Isolated DNA, as low as 20 ng, is enzymatically fragmented to generate small pieces of dsDNA. This is followed by the library construction step, in which IL-N7 adapters, molecular barcodes, and sample indexes are incorporated into DNA fragments generated in the previous step. Library fragments now serve as templates for target enrichment using single primer extension. In this step, targets are enriched using a single gene-specific primer and a universal forward primer. The final step is library amplification and sample indexing (for dual indexing) using the IL-S5 sample index primer and a universal primer.