QIAquick 96 PCR Purification Kit
96孔板纯化96个PCR产物(多至10 μg),100 bp–10 kb
96孔板纯化96个PCR产物(多至10 μg),100 bp–10 kb
✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
Cat. No. / ID: 28181
✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
QIAquick 96 PCR Purification Kits含有96孔板、缓冲液和收集管,利用硅胶模纯化技术高通量纯化大于100 bp PCR产物。通过简单、快速的结合、洗涤、洗脱的流程纯化长达10 kb的DNA,洗脱体积为60–80 μl(最终洗脱体积为40–60 μl)。回收过程可在BioRobot工作站上使用QIAquick 96 PCR BioRobot Kit全自动运行。
QIAquick 96 Kits包含一个硅胶膜,可在高盐缓冲液中结合DNA,并且在低盐缓冲液或水中洗脱。纯化过程去除引物、核苷酸、酶、矿物油、盐、琼脂糖、溴化乙锭和DNA样本中的其他杂质。硅胶膜技术结局了与树脂松动的问题和不便。优化特殊的结合缓冲液用于特殊应用,并且在特定的片段范围内,促进DNA的选择性吸附。
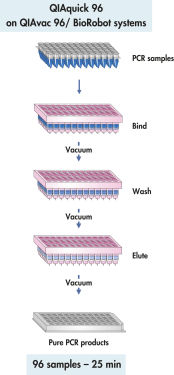
QIAquick 96操作流程平行纯化多达96个样本,并在QIAvac 96上采用高效真空驱动的纯化过程。
QIAquick体系采用简单的结合、洗涤、洗脱操作流程(参见"QIAquick 96 procedure")。将结合缓冲液直接加到PCR样本或者其他酶试剂中,然后将混合液上样到96孔板上。在高盐的缓冲液中,核酸吸附到硅胶膜上。洗去杂质,并且用少量的低盐缓冲液或纯水洗脱纯DNA,为后续应用准备。
QIAquick多孔模块在QIAvac底座上采用真空驱动的纯化过程处理。QIAquick 96 PCR Purification Kit需要使用QIAvac 96真空底座。产物回收过程可以使用QIAquick 96 PCR BioRobot Kit在BioRobot工作站上自动运行。
采用MinElute或者QIAquick体系纯化的DNA片段可即用于多种下游应用,包括测序、微阵列分析、连接和转化、限制性酶切、标记、显微注射、PCR和体外转录。
QIAquick 96 Kit 使用 QIAvac 96 或 BioRobot 通用系统进行结合-洗涤-洗脱。
| Features | Specifications |
|---|---|
| Binding capacity | 10 µg |
| Processing | 手动/自动 |
| Removal <10mers 17–40mers dye terminator proteins | 去除 <40mer |
| Sample type: applications | DNA、寡核苷酸:PCR 反应 |
| Format | 96 孔板 |
| Fragment size | 100 bp – 10 kb |
| Technology | 硅胶膜技术 |
| Recovery: oligonucleotides dsDNA | 回收率:寡核苷酸、dsDNA |
| Elution volume | 60-80 µl |