在 GeneGlobe 配置
寻找或定制设计合适的靶标特异性检测和组合,以研究您感兴趣的生物靶标。
Microbial DNA qPCR Arrays
Cat. No. / ID: 330261
Array plate and master mix for detection of microbial species or genes
在 GeneGlobe 配置 要查看价格
Microbial DNA qPCR Arrays 旨在用于分子生物学应用。这些产品不能用于疾病诊断、预防和治疗。
在 GeneGlobe 配置
寻找或定制设计合适的靶标特异性检测和组合,以研究您感兴趣的生物靶标。
特点
- 可检测微生物物种、毒力因子基因、或抗生素耐药性基因
- 适用于多种类型样本,实验流程简单
- 含有内参,可确保获得可靠结果
产品详情
Microbial DNA qPCR Arrays含有多种引物、探针、试剂、对照等,用于鉴定和分析微生物物种、毒力因子基因、或抗生素耐药性基因,适用于多种类型样本。物种鉴定试剂可检测细菌的16S rRNA基因和真菌的核糖体RNA基因,每个芯片都含有宿主DNA对照、细菌DNA对照、和监测PCR反应是否成功的对照。该芯片试剂盒中还提供Microbial qPCR Mastermix。该产品规格便利、操作简单,可用于各种实验室常用real-time PCR仪,实现对微生物物种和样本中基因的可靠鉴定和分析。
绩效
线性扩增与动态范围
Microbial DNA qPCR Assays可对10至106个拷贝的DNA模板进行符合现行规律的扩增(参见 Linearity and sensitivity of Microbial DNA qPCR Assays)。最低定量阈限(LLOQ)
LLOQ是指在标准曲线线性范围内的最低模板浓度(参见 Limit of detection versus lower limit of quantification)。所有Microbial DNA qPCR Assays中的93%具有低至100个基因拷贝的LLOQ(参见 The LLOQ for all Microbial DNA qPCR Assays reveals high sensitivity)。该系列产品中的92%微生物鉴定产品能达到这一LLOQ,95%的毒力因子检测产品和97%的抗生素耐药性基因检测产品能达到这一LLOQ(参见 The LLOQ for microbial identification Microbial DNA qPCR Assays reveals high sensitivity、 The LLOQ for virulence factor gene detection Microbial DNA qPCR Assays reveals high sensitivity和 The LLOQ for antibiotic resistance gene detection Microbial DNA qPCR Assays reveals high sensitivity)。特异性
所有Microbial DNA qPCR Assay都经过严格检验,确保能够灵敏检测某一物种或基因(参见 Microbial DNA qPCR Assays are highly specific)。对于能够检测一种以上五种或基因的产品,产品说明页上附有电脑预测可检测靶标的列表。即便样本中存在多种物种,该试剂盒仍有极高的检测特异性,此类样本包括粪便、痰和菌斑(参见 Microbial DNA qPCR Assays display high sensitivity even in complex metagenomic samples);其特异性可通过测序验证(参见 Specificity of the Antibiotic Resistance Genes Microbial DNA qPCR Array is confirmed by pyrosequencing)。
可重复性
同一实验员和不同实验员使用Microbial DNA qPCR Assays进行多次检测,都具有可重复性(参见 Microbial DNA qPCR Arrays generate reliably reproducible results)。查看图表
Linearity and sensitivity of Microbial DNA qPCR Arrays. Limit of detection versus lower limit of quantification.
Limit of detection versus lower limit of quantification.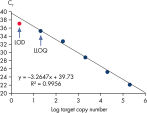 The lower limit of quantification (LLOQ) for all Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for all Microbial DNA qPCR Assays reveals high sensitivity.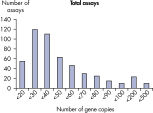 The lower limit of quantification (LLOQ) for microbial identification Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for microbial identification Microbial DNA qPCR Assays reveals high sensitivity.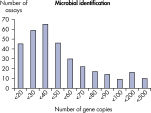 The lower limit of quantification (LLOQ) for virulence factor gene detection Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for virulence factor gene detection Microbial DNA qPCR Assays reveals high sensitivity.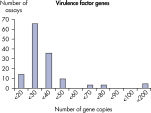 The lower limit of quantification (LLOQ) for antibiotic resistance gene detection Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for antibiotic resistance gene detection Microbial DNA qPCR Assays reveals high sensitivity.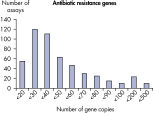 Microbial DNA qPCR Assays are highly specific.
Microbial DNA qPCR Assays are highly specific.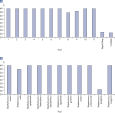 Microbial DNA qPCR Assays display high sensitivity even in complex metagenomic samples.
Microbial DNA qPCR Assays display high sensitivity even in complex metagenomic samples.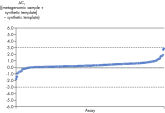 Specificity of the Antibiotic Resistance Genes Microbial DNA qPCR Array is verified by pyrosequencing.
Specificity of the Antibiotic Resistance Genes Microbial DNA qPCR Array is verified by pyrosequencing. Microbial DNA qPCR Arrays generate reliably reproducible results.
Microbial DNA qPCR Arrays generate reliably reproducible results.
 Limit of detection versus lower limit of quantification.
Limit of detection versus lower limit of quantification. The lower limit of quantification (LLOQ) for all Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for all Microbial DNA qPCR Assays reveals high sensitivity. The lower limit of quantification (LLOQ) for microbial identification Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for microbial identification Microbial DNA qPCR Assays reveals high sensitivity. The lower limit of quantification (LLOQ) for virulence factor gene detection Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for virulence factor gene detection Microbial DNA qPCR Assays reveals high sensitivity. The lower limit of quantification (LLOQ) for antibiotic resistance gene detection Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for antibiotic resistance gene detection Microbial DNA qPCR Assays reveals high sensitivity. Microbial DNA qPCR Assays are highly specific.
Microbial DNA qPCR Assays are highly specific. Microbial DNA qPCR Assays display high sensitivity even in complex metagenomic samples.
Microbial DNA qPCR Assays display high sensitivity even in complex metagenomic samples. Specificity of the Antibiotic Resistance Genes Microbial DNA qPCR Array is verified by pyrosequencing.
Specificity of the Antibiotic Resistance Genes Microbial DNA qPCR Array is verified by pyrosequencing. Microbial DNA qPCR Arrays generate reliably reproducible results.
Microbial DNA qPCR Arrays generate reliably reproducible results.
原理
Microbial DNA qPCR Assays在进行物种检测时,可检测细菌的16S rRNA基因和真菌核糖体rRNA基因序列;采用PCR扩增引物和水解探针,检测毒力因子基因和抗生素耐药性基因。
Microbial DNA qPCR Arrays为96孔板或384孔板规格,含有相应的引物、探针、对照和试剂等;例如,与呼吸道感染相关的微生物物种、或生物反恐研究的微生物等。每个孔板都含有多种对照,用于检测是否存在真菌DNA、细菌DNA、宿主基因组DNA,以及监测PCR反应是否成功,确保获得可靠结果。
Microbial DNA qPCR Arrays为96孔板或384孔板规格,含有相应的引物、探针、对照和试剂等;例如,与呼吸道感染相关的微生物物种、或生物反恐研究的微生物等。每个孔板都含有多种对照,用于检测是否存在真菌DNA、细菌DNA、宿主基因组DNA,以及监测PCR反应是否成功,确保获得可靠结果。
程序
Microbial DNA qPCR Assay的实验流程简单,能够在各种实验室real-time PCR仪上使用。使用适用于该类型样本的QIAamp试剂盒从样本中分离DNA,然后使用与PCR仪相应的Microbial qPCR Mastermix构建PCR反应体系。将混合液等分后加入孔板各孔,然后进行real-time PCR,获得每孔的原始CT值。然后使用免费的数据分析软件,分析样本中的基因或微生物物种。
应用
Microbial DNA qPCR Arrays用于鉴定和分析微生物物种或基因,适用于多种类型样本。例如,Vaginal Flora Microbial DNA qPCR Array可用于研究细菌性阴道炎的致病原因(参见 The Vaginal Flora Microbial DNA qPCR Array provides accurate profiling for cervical swab samples和 Vaginal samples positive for Gardnerella vaginalis also show changes in commensal and bacterial vaginosis-related microbes compared to healthy samples)。而Antibiotic Resistance Genes Microbial DNA qPCR Array可分析肠道或污水样本中的抗生素耐药性基因(参见 The Microbial DNA qPCR Array screens gut microbiota for the presence of antibiotic resistance genes和 The Antibiotic Resistance Genes Microbial DNA qPCR Array identified antibiotic resistance genes in sewage samples)。
查看图表
The Vaginal Flora Microbial DNA qPCR Array provides accurate profiling for cervical swab samples. Vaginal samples positive for Gardnerella vaginalis also show changes in commensal and bacterial vaginosis-related microbes compared to healthy samples.
Vaginal samples positive for Gardnerella vaginalis also show changes in commensal and bacterial vaginosis-related microbes compared to healthy samples.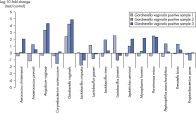 The Microbial DNA qPCR Array screens gut microbiota for the presence of antibiotic resistance genes.
The Microbial DNA qPCR Array screens gut microbiota for the presence of antibiotic resistance genes. The Antibiotic Resistance Genes Microbial qPCR Array identified antibiotic resistance genes in sewage samples.
The Antibiotic Resistance Genes Microbial qPCR Array identified antibiotic resistance genes in sewage samples.
 Vaginal samples positive for Gardnerella vaginalis also show changes in commensal and bacterial vaginosis-related microbes compared to healthy samples.
Vaginal samples positive for Gardnerella vaginalis also show changes in commensal and bacterial vaginosis-related microbes compared to healthy samples. The Microbial DNA qPCR Array screens gut microbiota for the presence of antibiotic resistance genes.
The Microbial DNA qPCR Array screens gut microbiota for the presence of antibiotic resistance genes. The Antibiotic Resistance Genes Microbial qPCR Array identified antibiotic resistance genes in sewage samples.
The Antibiotic Resistance Genes Microbial qPCR Array identified antibiotic resistance genes in sewage samples.
辅助数据和图表
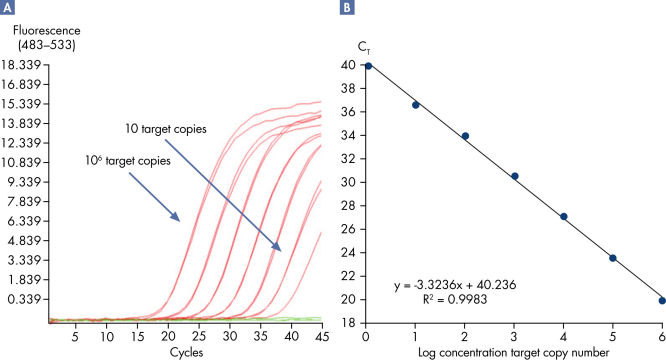
Linearity and sensitivity of Microbial DNA qPCR Arrays.
Linearity and sensitivity for each Microbial DNA qPCR Array was determined using synthetic templates over a 6 log serial dilution ranging from 1 copy to 1 million copies. The following are representative results for all the qPCR assays. [A] shows the real-time amplification curves of the KPC antibiotic resistance gene qPCR assay. In [B], a standard curve was prepared that shows that the primer efficiency equals 103% (calculated from slope = –3.3236) and the correlation coefficient is 0.9983, indicating optimum performance for the KPC qPCR assay. All Microbial DNA qPCR Assays have primer efficiencies between 80–120% and correlation coefficients (R)>0.995.
资源
下载文件 (20)
安全数据表 (1)
补充实验方案 (4)
在线讲座 (1)
分析软件 (1)
产品介绍与指南 (2)
学术海报 (1)
技术资讯 (2)
试剂盒操作手册 (1)
Safety Data Sheets (1)
Certificates of Analysis (1)
Brochures & Guides (2)
Download Files (20)
Analysis Software (1)
Kit Handbooks (1)
Webinars (1)
Scientific Posters (1)
Supplementary Protocols (4)
Technical Information (2)
FAQ
What species are detected by the Pan Bacteria 1 and Pan Bacteria 3 Assays?
What sequences are used to design the Microbial DNA qPCR Assays?
What are the storage conditions for the Microbial DNA qPCR products?
What is the difference between LLOQ and LOD?
What is the difference between Positive PCR Control (PPC) and Microbial DNA Positive Control?
Is the Microbial qPCR mastermix used in the Microbial DNA assay and in the Microbial DNA arrays free of genomic bacterial DNA?
What sample types can be tested on the arrays/assays?
Can I measure antibiotic resistance gene expression?
What is the expected amplicon size of the Microbial DNA qPCR Assays?
What is LLOQ?
Can I measure virulence factor gene expression?
Can I use the Microbial DNA-Free Water and Microbial qPCR Mastermix if they have been opened more than 3 times?
How can I calculate the number of bacterial cells that are present in a sample using the Microbial DNA qPCR Assays?
Are the Microbial DNA qPCR Assays wet-lab verified?
Are the assays species-specific?
Which Microbial qPCR Mastermix should I use?
What are the minimum sample requirements for Microbial DNA qPCR kits?
Which probe labels are available for the Microbial DNA qPCR Assays?
What is the sensitivity for the Microbial DNA qPCR kits?

