✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
QIAEX II Gel Extraction Kit (150)
Cat. No. / ID: 20021
✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
特点
- 有效提取 40 bp 至 50 kb 的 DNA
- 从 TAE 或 TBE 琼脂糖凝胶和聚丙烯酰胺凝胶中进行凝胶提取
- 无碘化钠干扰后续反应
- 不会剪切大的 DNA 片段
产品详情
QIAEX II 系统提供了一种二氧化硅颗粒悬浮液,在存在离液盐的情况下,DNA 片段与之结合。将 QIAEX II 悬浮液加入溶液或溶解的琼脂糖凝胶切片中,使其与 DNA 结合。通过短暂离心收集颗粒,进行清洗,然后用 Tris 缓冲液或水洗脱 40 bp 至 50 kb 的 DNA。
绩效
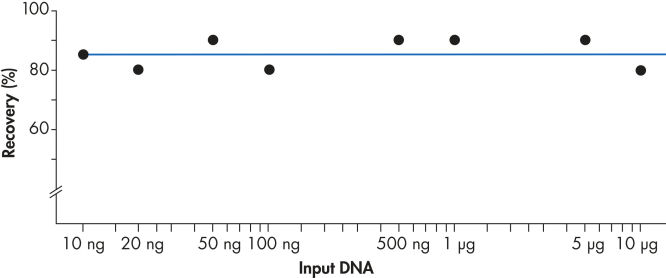
使用 QIAEX II 系统可有效回收 10 ng 至 10µg DNA(见图 “稳定的回收率”)。使用 30µl QIAEX II 悬浮液,可轻松将用于批量纯化凝胶片段的通用程序的结合能力放大到 15µg。
QIAEX II 系统提供的硅胶颗粒可纯化 60–95% 的 DNA 片段 (40 bp – 50 kb)。体积为 10µl 的 QIAEX II 悬浮液可结合多达 5µg 的 DNA,随后用 20µl 洗脱。
根据大小进行回收
| DNA 大小 | 回收率,百分比* |
|---|---|
| 44 bp | 75 |
| 75 bp | 75 |
| 500 bp | 95 |
| 7.5 kb | 85 |
| 23.5 kb | 75 |
| 48.5 kb | 60 |
查看图表
原理
使用 QIAEX II 系统纯化 DNA 片段的基础是琼脂糖的溶解和核酸在离液盐存在的情况下对 QIAEX II 硅胶颗粒的选择性吸附。QIAEX II 能从盐类、琼脂糖、聚丙烯酰胺、染料、蛋白质和核苷酸中分离出 DNA,且无需苯酚提取或乙醇沉淀。QIAEX II 对 TAE 或 TBE 缓冲液中任何类型的琼脂糖都有效。
QIAEX II 颗粒可为凝胶提取提供稀浆,确保即使是大的 DNA 片段也能在不剪切的情况下高效回收。经过优化的缓冲液可以在不使用碘化钠的情况下回收 DNA,而碘化钠是很难从 DNA 样本中去除的,并且还可能会影响后续反应。
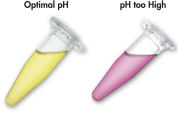
QIAEX II 系统使用的溶解和结合缓冲液含有独特的 pH 指示剂。简单的颜色变化就能显示结合混合物的 pH 值是否最适合 QIAEX II 硅胶颗粒对 DNA 的有效吸附(见图 “pH 指示基团染料”)。有颜色的染料还能让人轻松看到结合混合物中任何未溶解的琼脂糖,确保完全溶解以获得最大产量。
借助溶解和结合缓冲液中的 pH 指示基团染料,可以方便地通过目测确定 DNA 吸附的最佳 pH 值 (pH ≤7.5)。如果琼脂糖凝胶电泳缓冲液使用频繁或配制不正确,会导致结合混合物 pH 值不正确。在这种情况下,只需加入 10 µl 3 M 乙酸钠(pH 值为 5.0)即可轻松调节 pH 值。
查看图表
程序
将 QIAEX II 硅胶颗粒加入已溶解的凝胶切片中,通过短暂的离心步骤收集颗粒(见流程图 “QIAEX II 程序”)。洗涤后,用 20 µl Tris 缓冲液或水洗脱纯 DNA 片段。
QIAEX II 系统提供 QIAEX II 悬浮液、结合缓冲液、洗涤缓冲液和一本全面的手册。提供了从琼脂糖凝胶、溶液和聚丙烯酰胺凝胶中纯化 DNA 的方案。
查看图表
应用
用 QIAEX II 系统纯化的 DNA 可直接用于大多数应用,包括
- 限制性酶切
- 标记
- 连接
- PCR
| 特点 | 规格 |
|---|---|
| 结合能力 | 5 µg/10 µl |
| 洗脱体积 | 20 µl |
| 格式 | 试管 |
| 片段大小 | 40 bp – 50 kb |
| 处理 | 手动 |
| 回收率:寡核苷酸 dsDNA | 回收率:dsDNA 片段 |
| 去除 <10mer 17–40mer 染料终止物蛋白质 | 去除 <40mer |
| 样本类型:应用 | DNA:PCR 反应 |
| 技术 | 硅胶膜技术 |
辅助数据和图表
稳定的回收率。