✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
QIAquick Nucleotide Removal Kit (250)
Cat. No. / ID: 28306
✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
特点
- 即用型 DNA 的回收率高达 95%
- 快速便捷的程序
- 只需三个简单步骤,即可净化长达 10 kb 的 DNA
- 凝胶加载染料方便样本分析
产品详情
QIAquick Nucleotide Removal Kit 提供离心柱、缓冲液和采样管,用于以硅胶膜为基础纯化寡核苷酸和 DNA。该试剂盒可去除未结合的核苷酸、盐和其他杂质,并通过简单快速的洗脱程序和 30–200 µl 的洗脱体积纯化大小 40 bp 到 10 kb 的寡核苷酸 (>17 nt) 和 DNA 片段。程序在 QIAcube Connect 上全自动完成。
绩效
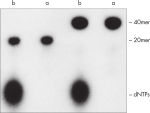
QIAquick 核苷酸去除程序可去除 DNA 样本中的 核苷酸、酶、盐和其他杂质(见图“ 完全去除标记寡核苷酸中的核苷酸”)。使用微型离心机纯化 17mer – 10 kb 的 DNA。如果样本量小于 50 µl,也可使用 DyeEx Spin 试剂盒。
查看图表
原理
QIAquick Kit 包含一个硅胶膜组件,用于在高盐缓冲液中结合 DNA,然后用低盐缓冲液或水进行洗脱。纯化程序可去除 DNA 样本中的引物、核苷酸、酶、矿物油、盐、琼脂糖、溴化乙锭和其他杂质(见图“ 完全去除标记寡核苷酸中的核苷酸”)。硅胶膜技术消除了树脂松散和稀浆带来的问题和不便。专用结合缓冲液针对特定应用进行了优化,可促进特定大小范围内 DNA 分子的选择性吸附。
凝胶加载染料
为了更快、更方便地进行样本处理和分析,我们提供了凝胶加载染料。GelPilot Loading Dye 包含三种跟踪染料(二甲苯胍青、溴酚蓝和橙黄 G),便于优化琼脂糖凝胶的运行时间,防止较小的 DNA 片段迁移过远(见图“ GelPilot Loading Dye”)。
查看图表
程序
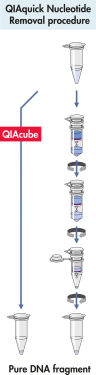
QIAquick 系统使用简单的结合–洗涤–洗脱程序(见流程图“ QIAquick 核苷酸去除程序”)。将结合缓冲液直接加到样本中,然后将混合物加到 QIAquick 离心柱上。核酸在缓冲液提供的高盐条件下吸附到硅胶膜上。杂质被洗去,用少量低盐缓冲液或水洗脱出的纯 DNA 可直接用于所有后续应用。
处理
QIAquick 离心柱的设计提供了两种方便的处理选项。离心柱可放入传统的台式微型离心机。QIAquick Nucleotide Removal Kit 以及其他基于 QIAGEN 离心柱的试剂盒均可在 QIAcube Connect 上实现全自动操作,从而提高生产效率和结果的规范化(见图“离心柱处理选项 A”和“ QIAcube Connect”)。
查看图表
应用
用 QIAquick 系统纯化的 DNA 片段可直接用于所有应用,包括测序、微阵列分析、连接和转化、限制性酶切、标记和微量注射。
辅助数据和图表
QIAquick 核苷酸去除程序。
Specifications
| Features | Specifications |
|---|---|
| Binding capacity | 10 µg |
| Technology | 硅胶膜技术 |
| Recovery: oligonucleotides dsDNA | 回收率:寡核苷酸、dsDNA |
| Format | 试管 |
| Elution volume | 30-50 µl |
| Processing | 手动 |
| Sample type: applications | DNA、寡核苷酸:PCR 反应 |
| Removal <10mers 17–40mers dye terminator proteins | 去除 <10mer |
| Fragment size | 40 bp – 10 kb |