dPCR Copy Number Assay (200)
Cat. No. / ID: 250205
Features
- Predesigned assays for all genes in the human genome
- Choice of 3 design locations per gene: 5’, middle and 3’
- Even more predesigned assays available on request
- Assays for >200 targets have been dPCR wet-lab tested
- Simple and straightforward format for dPCR use
- Convenient copy number data analysis using the QIAcuity Software Suite
Product Details
dPCR Copy Number Assays enable specific, accurate, reproducible and easy-to-interpret copy number change analysis for an individual gene or region of interest. Assays for more than 200 targets have been dPCR wet-lab tested. All other in silico designs have been bench-verified and are ready to use in NGS follow-up studies, specific target screening and other related studies.
The dPCR Copy Number Assays are available in tube format as a ready-to-use 25x-concentrated assay. The tube contains the primer pair to be used with the QIAcuity EG PCR Kit and the QIAcuity instrument. dPCR wet-lab tested assays are flagged as such in the specific assay product view.
Performance
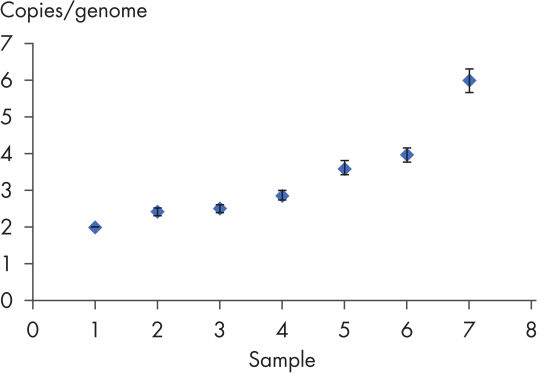
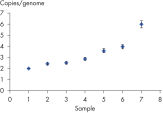
Laboratory verification of assay quality ensures that the dPCR Copy Number PCR Assays deliver reliable results. Copy number analysis by digital PCR can accurately identify aneuploidy in cell lines containing chromosomal aberrations previously identified by cytogenetic methods. Digital PCR is able to detect changes in copy number as small as 1.2 fold or from 5 to 6 copies. The partitioning of the dPCR reaction is the key factor that enables the level of discrimination required to resolve such small changes.
For normalization, Reference Assays corresponding to different copy numbers per human genome are also available.
Principle
All dPCR Copy Number PCR Assays are designed for unique regions of the human genome. The dPCR Copy Number Reference Assays (available separately) include a variety of references with different numbers of copies in the genome, making it possible to select the optimal normalization references for each analysis. We recommend including two reference assays to allow accurate calculation of the copy number of the gene or target of interest.
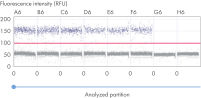
The dPCR Copy Number Assays are ready to use: just add the DNA sample and mastermix from the QIAcuity EG PCR Kit to the individual assays or reference assays in the QIAcuity Nanoplate. Detection is based on the intercalating dye EvaGreen.
The principle of the dPCR reaction in the nanoplates is described here.
Procedure
To set up the dPCR experiment, add an aliquot of the genomic DNA sample (isolated from fresh, frozen or fixed samples) to the QIAcuity EG PCR Kit mastermix and the dPCR Copy Number Assay. Each gene or target of interest and each reference assay is analyzed in its own individual well.
After loading and sealing the nanoplate, run the plate with the recommended cycling program on the QIAcuity instrument. The resulting copies per microliter are shown in the absolute quantification analysis type of the QIAcuity Software Suite. For the calculation of copy number per genome, select the copy number data analysis type in the QIAcuity Software Suite, and define the nanoplate wells for the gene/region of interest or the normalization reference.
Applications
dPCR Copy Number Assays are highly suited for accurately detecting copy number alterations or variations at individual loci using DNA from fresh, frozen or fixed samples.
Supporting data and figures
MYC copy number determination in MCF-7 cell line.