QIAamp DNA FFPE Advanced UNG Kit (50)
Cat. No. / ID: 56704
Features
- High recovery of amplifiable DNA
- Paraffin removal without xylene or similar solvents
- Uracil (deaminated cytosine)–artifact removal step using uracil-N-glycosylase (UNG) during DNA extraction
- Ready-to-use DNA for PCR, digital PCR (dPCR) and next-generation sequencing (NGS)
Product Details
Increase your recovery of high-quality DNA from FFPE tissue with the QIAamp DNA FFPE Advanced Kits’ xylene-free, no-wash deparaffinization, double-lyse protocol, and UCP (ultra-clean production) spin column technology.
Remove C→U transitions of nucleic acids with the kits’ optional UNG digest to optimize your recovered DNA for NGS analysis.
You can also automate the QIAamp DNA FFPE Advanced protocols on the QIAcube Connect.>
Performance
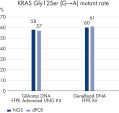
One characteristic of FFPE DNA quantification is that different methods will give out different results, and high UV-vis or fluorometric values do not necessarily mean good PCR performance.
Because real-time PCR, or quantitative PCR (qPCR), is one of the most common downstream applications for FFPE, the QIAamp DNA FFPE Advanced Kits are optimized for qPCR.
Regardless of values obtained in UV-vis or fluorometric measurements, QIAamp DNA FFPE Advanced Kits consistently showed better PCR performance under qPCR quantification than the other products tested (see figure “ Optimized for PCR performance”).
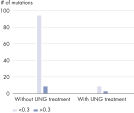
The QIAamp DNA FFPE Advanced UNG Kit is also well suited for purifying DNA to be used in NGS analysis, because it addresses the issue of artificial C→T/G→A transitions, which commonly occur in FFPE material due to cytosine deamination (see “ Artificial C→T/G→A transitions”).
Through UNG-based uracil digestion during DNA isolation, the QIAamp DNA FFPE Advanced UNG protocol reduces false-positive reports of single-nucleotide variants (SNVs) in NGS (see figure “ Dramatic reduction in artifactual C→T | G→A transitions”).
See figures
Principle
There are three major challenges in preparing DNA from FFPE tissues:
- Low yields due to limited input material and compromised DNA
- Nonamplifiable DNA due to formalin-induced crosslinks
- Deaminated-cytosine artifacts can cause false results in NGS for mutational analyses
The QIAamp DNA FFPE Advanced Kits maximize DNA yields from limited sample inputs in two ways:
- By implementing a two-step lysis procedure, which ensures high DNA extraction even from difficult-to-lyse samples
- By replacing solvent-based paraffin removal with the use of Deparaffinization Solution, which eliminates all wash steps before initial lysis, thus minimizing the risk of losing scarce sample material
Crosslink removal further increases the recovery of amplifiable DNA (see figure “ Optimized for PCR performance”).
Optional UNG treatment before the second lysis removes deaminated-cytosine artifacts, making the DNA especially suitable for NGS analysis (see table “ Primed for NGS”, as well as figures “ Reliable dPCR and NGS results” and “ Dramatic reduction in artifactual C→T | G→A transitions”).
See figures
Procedure
The QIAamp DNA FFPE Advanced procedure consists of a simplified deparaffinization step, two lysis steps with in-between de-crosslinking and optional artifact removal, and the standard bind-wash-elute steps (see figure “ QIAamp DNA FFPE Advanced workflow”).
See figures
Applications
DNA from FFPE using the QIAamp DNA FFPE Advanced Kits can be used immediately for PCR, dPCR or NGS, or it can be stored at −30 to −15°C.
Supporting data and figures
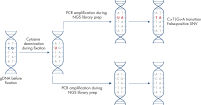
QIAamp DNA FFPE Advanced workflow.
The QIAamp DNA FFPE Advanced procedure removes paraffin without the tediousness of multiple wash steps, by using Deparaffinization Solution instead of xylene or any other solvent.
Lysis is done with Proteinase K. Cross-links are removed by heat incubation at 90°C for 1 hour. Artifacts can be removed with UNG.
RNA is then digested, and the sample is lysed a second time to increase DNA recovery.
DNA is bound with the QIAamp UCP MinElute spin column. Contaminants are washed off using Buffer AW1, Buffer AW2 and ethanol, and DNA is eluted in Buffer ATE.

Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, digital PCR, next-generation sequencing |
| Elution volume | 20–100 µl |
| Format | Spin column |
| Main sample type | Formalin-fixed paraffin-embedded (FFPE) tissue samples |
| Processing | Manual or automated with the QIAcube Connect |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein | Genomic DNA |
| Sample amount | Tissue sections, each with a thickness of 5–10 µm, for a total volume of 4 mm3 |
| Technology | Silica technology |