✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
EpiTect MethyLight PCR +ROX Vial Kit (200)
Cat. No. / ID: 59496
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Highly accurate TaqMan® probe-based real-time analysis
- Reliable methylation quantification with MethyLight Assays
- Convenient master mix format
Product Details
Performance
See figures
Principle
EpiTect MethyLight PCR Kits can be used in combination with EpiTect MethyLight Assays or other dual-labeled assays for sensitive quantification of DNA methylation in bisulfite converted DNA. Depending on the level used for sequence discrimination, MethyLight analyses can be performed in a quantitative or semiquantitative format.
Dual-labeled probes, including TaqMan® probes, are sequence-specific oligonucleotides with a fluorophore and a quencher moiety attached. The fluorophore is at the 5' end of the probe, and the quencher moiety is usually located internally or at the 3' end. During the elongation phase of PCR, the probe is cleaved by the 5' → 3' exonuclease activity the polymerase, separating the fluorophore from the quencher moiety. This results in detectable fluorescence that is proportional to the amount of accumulated PCR product.
Quantitative MethyLight real-time PCR with methylation-specific probes
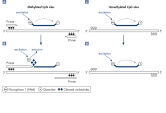
For quantitative methylation analysis using MethyLight PCR, two dual-labeled probes with different fluorescent labels are used in combination with one set of PCR primers. Bisulfite converted DNA requires a different probe sequence for methylated and unmethylated CpG sites. The primers are specific for unmethylated sites. One of the probes binds specifically to methylated DNA at the site of interest, and the other binds specifically to unmethylated DNA at the same site. Since the probes contain different fluorophores, the amounts of methylated and unmethylated sequence can be determined (see figure " Quantitative MethyLight real-time PCR").
Semiquantitative MethyLight real-time PCR with methylation-specific primers
For semiquantitative MethyLight analysis, a dual-labeled probe is used together with a set of PCR primers that specifically binds to the methylation sites of interest. Bisulfite converted DNA requires a different primer sequence for methylated and unmethylated CpG sites, so the primers need to be specific to methylated, bisulfite converted DNA. If the methylation-specific primer binds to the DNA, it will be elongated, and the 5' → 3' exonuclease activity of Taq DNA Polymerase will lead to the degradation of the primer and the release of the fluorophore. As the fluorophore is now separated from the quencher moiety, its fluorescence is detectable (see figure " Semiquantitative MethyLight real-time PCR").
Increased specificity
The EpiTect MethyLight Master Mix has been specifically developed for highly sensitive detection of methylated and unmethylated DNA using sequence-specific probes. In addition to various salts and additives, the buffer also contains a specially optimized combination of KCl and (NH4)2SO4, which promotes a high ratio of specific-to-nonspecific primer and probe binding during the annealing step of each PCR cycle. This allows discriminative hybridization of the probes, and the detection of unmethylated and/or methylated target sequences. The stringent primer annealing conditions lead to increased PCR specificity when amplifying bisulfite converted DNA, enabling reliable probe-based methylation detection.
Control reactions in MethyLight PCR
Control reactions must be carried out to ensure that MethyLight PCR probes and primers specifically bind methylated or unmethylated DNA. Such reactions require bisulfite converted control DNA (fully methylated and fully unmethylated) in various concentrations. In addition, mixtures of these control DNAs can serve as quantification standards when determining methylation degrees. With the EpiTect Control DNA Set, QIAGEN provides the quality-controlled DNAs needed — methylated bisulfite converted, unmethylated bisulfite converted, and normal human genomic DNA — in a ready-to-use kit format for standardized and reliable methylation-specific control reactions.
See figures
Procedure
The EpiTect MethyLight PCR Kit is supplied in a master mix format for a fast and convenient reaction setup. Two kits are available, with and without the inclusion of ROX in the master mix, offering greater flexibility for different real-time cyclers. Validated protocols for most real-time cyclers guarantee optimal results.
The EpiTect MethyLight PCR Kit is supplied with a master mix containing ROX passive reference dye, and is optimized for use with real-time cyclers that require a high concentration of ROX dye for fluorescence normalization (e.g., instruments from Applied Biosystems, excluding the Applied Biosystems 7500 Real-Time PCR Systems).
The EpiTect MethyLight PCR + ROX Vial Kit is supplied with a master mix that does not contain ROX dye, but it includes a separate solution of ROX dye that can be added to reactions, depending on the real-time cycler used. The kit is intended for cyclers that require a lower concentration of ROX dye for fluorescence normalization (e.g., Applied Biosystems 7500 Real-Time PCR Systems), for use with cyclers that allow optional use of ROX dye (e.g., instruments from Stratagene), and for use with cyclers that do not require ROX dye.
Standardized workflows in epigenetics
Accessing epigenetic information is of prime importance for many areas of biological and medical research — particularly oncology, but also stem cell research and developmental biology. However, the analysis of changes in DNA methylation is challenging due to the lack of standardized methods for providing reproducible data particularly from limited sample material. With its newly introduced EpiTect solutions, QIAGEN makes available standardized, pre-analytical and analytical solutions, from DNA sample collection, stabilization and purification, to bisulfite conversion and real-time or end-point PCR methylation analysis or sequencing (see figure " Standardized workflows in epigenetics").
See figures
Applications
EpiTect MethyLight PCR Kits are suitable for quantitative and semiquantitative real-time PCR analysis and can be used in combination with EpiTect MethyLight Assays or other dual-labeled assays.
Supporting data and figures
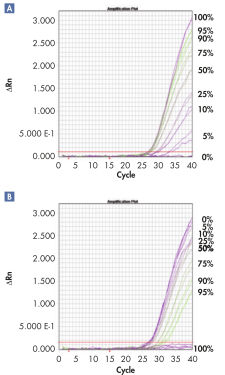
Sensitive real-time PCR quantification.
Specifications
| Features | Specifications |
|---|---|
| Applications | Quantification of bisulfite converted DNA |
| Reaction type | Highly accurate and sensitive probe-based MethyLight real-time analysis |
| Real-time or endpoint | Real-time |
| With or without ROX | Available with ROX in master mix and with ROX as separate vial |
| Sample/target type | Bisulfite converted DNA |
| SYBR Green I or sequence-specific probes | sequence-specific probes |
| Single or multiplex | Single & duplex |
| Thermal cycler | Most real-time cyclers except Applied Biosystems 7500 systems |