✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
EpiTect Bisulfite Kit (48)
Cat. No. / ID: 59104
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Streamlined procedure for fast and reliable results
- Complete cytosine conversion
- Unique DNA protection system for highly sensitive analysis
- Optimized protocols, including conversion from FFPE DNA
- Spin-column and 96-well plate formats for high throughput
Product Details
EpiTect Bisulfite Kits enable complete conversion of unmethylated cytosines to uracils and subsequent purification in 6 hours. The EpiTect 96 Bisulfite procedure takes less than 7 hours. The highly sensitive method utilizes innovative protection against DNA degradation that guarantees sensitive results, even from 1 ng DNA, and ensures high conversion rates of over 99%. Due to its unique DNA Protect technology, EpiTect Bisulfite converted DNA is highly suitable for whole bisulfitome amplification using EpiTect Whole Bisulfitome Kits. This enables reliable amplification of DNA in cases where bisulfite converted DNA is limited. EpiTect Bisulfite Kits include a spin-column or 96-well format procedure, and can be processed using a centrifuge or vacuum manifold. In addition, the EpiTect Bisulfite Kit can be fully automated on the QIAcube Connect.
Performance
Rapid results
The complete EpiTect Bisulfite Kit centrifugation or vacuum procedure — from DNA to pure bisulfite converted DNA ready for analysis — takes only 6 hours. The EpiTect 96 Bisulfite procedure takes under 7 hours. Traditional bisulfite conversion procedures can take over 18 hours and require extensive user interaction (see table "Fast and Easy Bisulfite Conversion" and figure " Time saving procedure"). The EpiTect Bisulfite Kit provides everything required for successful bisulfite conversion and DNA cleanup.
| EpiTect procedure | Traditional procedure | |
| Reagent setup | 10 minutes; prealiquoted solutions | 40 minutes; reagent setup needed |
| Bisulfite reaction | 5 hours | 16 hours |
| Purification | 30–45 minutes; EpiTect spin columns and plates | 120 minutes; purification, desulfonation, precipitation, and washing steps |
Complete DNA conversion
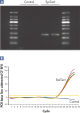
Each EpiTect Bisulfite Kit reaction converts 1 ng – 2 µg of DNA with equal efficiency. The novel procedure and optimized buffers enable the recovery of high yields of converted DNA. The highly efficient cytosine conversion is apparent by the low CT values obtained when amplifying converted DNA using real-time PCR (see figure " Complete cytosine conversion"). Analysis of EpiTect Bisulfite Kit converted DNA shows over 99% conversion of unmethylated cytosines (see figure " Highly efficient cytosine conversion"). This high conversion rate provides repeatable and dependable downstream analysis.
See figures
Principle
Bisulfite conversion of challenging samples
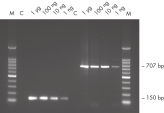
Due to the limited amount of DNA available, determination of methylation patterns in DNA from precious and limited sample materials (e.g., formalin-fixed paraffin-embedded [FFPE] or microdissected tissue) is especially challenging. However, such sample types are of specific interest for the successful identification of valuable disease-predicting biomarkers. EpiTect Bisulfite Kits address these challenges with optimized protocols for these sample types (see figure " Methylation detection from FFPE tissue samples"). All of the required buffers and reagents are provided, and only minimal handling is required.
For direct bisulfite conversion of DNA from FFPE samples, cells, blood, and tissue without prior DNA isolation, we recommend EpiTect Plus FFPE Bisulfite Kits and EpiTect Plus LyseAll Bisulfite Kits.
Preventing DNA fragmentation
DNA Protect Buffer is uniquely formulated to prevent DNA fragmentation (usually associated with bisulfite treatment of DNA at high temperatures and low pH values) and provide effective DNA denaturation, resulting in the single-stranded DNA necessary for complete cytosine conversion (see " Unique DNA Protect Buffer and pH indicator system"). The prevention of fragmentation enables subsequent amplification and analysis of large PCR fragments (see figure " Fast and easy bisulfite conversion"). DNA Protect Buffer also contains a pH indicator dye, allowing confirmation of the correct pH for cytosine conversion. The efficient integrated DNA cleanup allows storage of the converted DNA for at least 12 months without affecting the DNA quality (see figure " Long-term DNA storage").
See figures
Procedure
The EpiTect procedure comprises a few simple steps, based on QIAGEN's straightforward bind, wash, and elute system (see flowchart " Conversion procedure"). After thermal denaturation and sodium bisulfite DNA conversion, the DNA is applied to an EpiTect spin column or plate where, using optimized buffers and a standard microcentrifuge or vacuum manifold, it is washed to remove all traces of sodium bisulfite and eluted. The eluted DNA can be used in all downstream applications, including methylation-specific PCR, real-time PCR analysis, bisulfite sequencing, COBRA, and Pyrosequencing.
Accessing epigenetic information is of prime importance for many areas of biological and medical research — particularly oncology, but also stem cell research and developmental biology. However, the analysis of changes in DNA methylation is challenging due to the lack of standardized methods for providing reproducible data, particularly from limited sample material. With its newly introduced EpiTect solutions, QIAGEN makes available standardized, pre-analytical and analytical solutions: from DNA sample collection, stabilization and purification, to bisulfite conversion and real-time or end-point PCR methylation analysis or sequencing (see figure " Standardized workflows in epigenetics").
See figures
Applications
DNA converted and purified using the EpiTect Bisulfite Kit is suitable for use in a range of downstream applications, including:
- Methylation-specific PCR, real-time PCR
- COBRA
- Sequencing/Pyrosequencing/NGS
- High-resolution melting
- Methylation arrays
Supporting data and figures
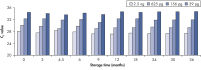
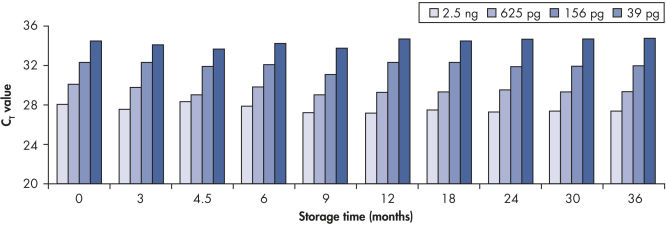
Long-term DNA storage.
Bisulfite converted DNA generated with EpiTect Kits was stored at –20°C for up to 36 months, and amplified at several time intervals. The resulting threshold cycle values at all time points for various DNA amounts were comparable to those obtained with freshly converted DNA (time point 0).
Specifications
| Features | Specifications |
|---|---|
| Applications | Multiplex PCR, real-time PCR, sequencing |
| Processing | Manual |
| Sample types | Blood, (FFPE) tissue, DNA samples |
| Conversion efficiency | 99.4–99.8% |
| Format | Spin column and 96-well plate |
| Time per run or per prep | <6 hours (1-48 samples), ~ 7 hours (48-96 samples) |
| Elution volume | Varies |
| Sample amount | 1–2 µg |
| Yield | Depends on input amount of purified genomic DNA used for conversion |