✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
QIAEX II Gel Extraction Kit (150)
Cat. No. / ID: 20021
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Efficient extraction of DNA from 40 bp to 50 kb
- Gel extraction from TAE or TBE agarose gels and polyacrylamide gels
- No sodium iodide to interfere with subsequent reactions
- No shearing of large DNA fragments
Product Details
The QIAEX II system provides a suspension of silica particles to which DNA fragments bind in the presence of chaotropic salts. QIAEX II Suspension is added to solutions or solubilized agarose gel slices and binds DNA. The particles are collected by a brief centrifugation, washed and DNA of 40 bp to 50 kb is eluted in Tris buffer or water.
Performance
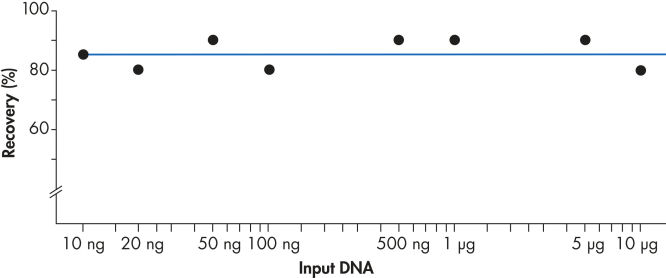
Using the QIAEX II system, 10 ng to 10 µg DNA are recovered efficiently (see figure "Consistent recovery"). The versatile procedure for batch purification of gel fragments can be easily scaled up to a binding capacity of 15 µg using 30 µl QIAEX II suspension.
The QIAEX II system provides silica particles for purifying 60–95% DNA fragments (40 bp – 50 kb). A volume of 10 µl QIAEX II suspension binds up to 5 µg DNA, which is subsequently eluted in 20 µl.
Recovery according to size
| DNA size | Recovery, percent* |
|---|---|
| 44 bp | 75 |
| 75 bp | 75 |
| 500 bp | 95 |
| 7.5 kb | 85 |
| 23.5 kb | 75 |
| 48.5 kb | 60 |
See figures
Principle
Purification of DNA fragments with the QIAEX II system is based on solubilization of agarose and selective adsorption of nucleic acids onto QIAEX II silica-gel particles in the presence of chaotropic salt. QIAEX II separates DNA from salts, agarose, polyacrylamide, dyes, proteins and nucleotides without phenol extraction or ethanol precipitation. QIAEX II is effective for any type of agarose in either TAE or TBE buffers.
QIAEX II particles provide a slurry for gel extraction and ensure efficient recovery without shearing, even for large DNA fragments. Optimized buffers permit DNA recovery without sodium iodide, which is difficult to remove from DNA samples, and may affect subsequent reactions.
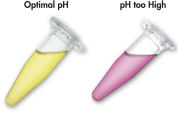
The solubilization and binding buffer used with the QIAEX II system contains a unique pH indicator. A simple color change indicates whether the pH of the binding mixture is optimal for efficient adsorption of DNA to QIAEX II silica particles (see figure "pH indicator dye"). The colored dye also allows easy visualization of any unsolubilized agarose in the binding mixture, ensuring complete solubilization for maximum yield.
pH indicator dye in the solubilization and binding buffer allows easy visual determination of optimal pH for DNA adsorption (pH ≤7.5). An incorrect binding-mixture pH can occur if the agarose gel electrophoresis buffer was frequently used or incorrectly prepared. In this case the pH can be easily adjusted by addition of 10 µl 3 M sodium acetate, pH 5.0.
See figures
Procedure
QIAEX II silica-gel particles are added to the solubilized gel slice, and the particles collected by a brief centrifugation step (see flowchart "QIAEX II procedure"). After washing, the pure DNA fragment is eluted in 20 µl of Tris buffer or water.
The QIAEX II system provides QIAEX II suspension together with binding and wash buffers, and a comprehensive handbook. Protocols are provided for purification of DNA from agarose gels, solutions and polyacrylamide gels.
See figures
Applications
DNA purified with the QIAEX II system can be used directly in most applications, including:
- Restriction digestion
- Labeling
- Ligation
- PCR
| Features | Specifications |
|---|---|
| Binding capacity | 5 µg/10 µl |
| Elution volume | 20 µl |
| Format | Tube |
| Fragment size | 40 bp – 50 kb |
| Processing | Manual |
| Recovery: oligonucleotides dsDNA | Recovery: dsDNA fragments |
| Removal <10mers 17–40mers dye terminator proteins | Removal <40mers |
| Sample type: applications | DNA: PCR reactions |
| Technology | Silica technology |
Supporting data and figures
Consistent recovery.