Products

SARS-CoV-2 Neo Assay Kit (600)
Cat. No. / ID: 222115

SARS-CoV-2 Neo Assay Kit (2400)
Cat. No. / ID: 222117

SARS-CoV-2 Neo Positive Control
Cat. No. / ID: 222710
Features
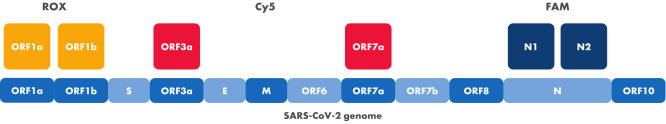
- Targets ORF1a, ORF1b, ORF3a, ORF7a, N1 and N2 within the SARS-CoV-2 genome
- Compatible with nasopharyngeal and oropharyngeal swabs, lolly (lollipop swab), gargle and saliva
- Detects six different targets in three color channels (FAM, ROX and Cy5)
- Does not detect SARS-CoV-1 or other Sarbecovirus species
- Ready-to-use assay mix (all-in-one tube)
Product Details
To ensure future-proof detection of SARS-CoV-2, we developed a novel assay kit covering six targets across four genomic regions (ORF1ab, ORF3a, ORF7a and N gene) of the virus. Using multiple targets in multiple regions increases the likelihood of successful detection.
Probes are coupled to different dyes and can be measured in the FAM, ROX and Cy5 channels on a cycler of your choice.
This kit can be used in combination with the QIAprep& Viral RNA UM Kit chemistry and SARS-CoV-2 Neo Positive Control to provide fast and easy detection. Sample types compatible for detecting SARS-CoV-2 include nasal, nasopharyngeal, oropharyngeal swabs, lolly (lollipop swab), saliva, and gargles.
Performance
The unique design of the SARS-CoV-2 Neo Assay enables reliable and highly specific SARS-CoV-2 detection. In silico analyses showed that all investigated SARS-CoV-2 viral sequences (>4.8 Mio) were successfully detected by this assay. The assay does not detect SARS-CoV-1, other Sarbecovirus species, and/or other organisms as demonstrated by wet-lab and in-silico analyses. The list of wet-lab-tested respiratory pathogens includes Influenza A (H3N2, H1N1), Influenza B, Parainfluenza (1-4), Coronavirus 229E, OC43, NL63, C. pneumoniae, Metapneumovirus, B. pertussis, Human Rhinovirus 1A, RSV A, Human Adenovirus 5, L. pneumophila and Bocavirus. RNA from different respiratory pathogen-positive controls (chemically inactivated viruses) was tested for cross-reactivity with the SARS-CoV-2 Neo Assay using the QIAprep& Viral RNA UM Kit. None of the pathogens examined showed cross-reactivity (data not shown).
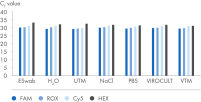
The SARS-CoV-2 Neo Assay Kit can be used in combination with the QIAprep& Viral RNA UM Kit chemistry and is compatible with cyclers having FAM, ROX and Cy5 detection channels and samples collected in all non-fixation transport media. Pooling of up to 20 samples per reaction is possible.
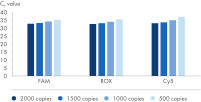
The analytical sensitivity per channel was assessed using in-vitro transcribed nucleic acids and determined as follows: 1000 copies/ml in FAM, 1000 copies/ml in ROX and 1500 copies/ml in Cy5.
Principle
The SARS-CoV-2 Neo Assay is intended for use with the QIAprep& Viral RNA UM Kit, with the above-mentioned sample types, transport media and cyclers.
The SARS-CoV-2 Neo Assay consists of twelve primers and six probes. Primers and probes are pre-mixed in a ready-to-use 20x formulation in a single tube. The oligonucleotides amplify the following six targets in the viral RNA: ORF1a, ORF1b, ORF3a, ORF7a, N1 and N2. The unique target combination results in high mutational tolerance while ensuring highly specific SARS-CoV-2 detection.
The probes are coupled to different dyes which can be detected in the FAM, ROX and Cy5 channels.
Optionally, the SARS-CoV-2 Neo Positive Control containing a synthetic DNA fragment covering all six targets can be used in the experimental setup.
Procedure
The SARS-CoV-2 Neo Assay is optimized for use with the QIAprep& Viral RNA UM Kit. Follow a few simple steps as outlined in the Quick-Start Protocol and get your results in under one hour. The procedure does not require a separate RNA extraction.
The SARS-CoV-2 Neo Assay uses the human sampling control of the QIAprep& Viral RNA UM Kit to monitor successful sampling and PCR amplification of the sample of your choice. You can use any qPCR instrument capable of detecting FAM, ROX, Cy5 and HEX (for the human sampling control).
The simple workflow is automatable on your platform of choice. If using a pool of 10 samples per reaction on a high-capacity robotic platform, a throughput of 250,000 samples per week can be achieved when working 24/7.
Applications
The SARS-CoV-2 Neo Assay Kit is intended for molecular biology applications such as epidemiological research using real-time RT-PCR.
Supporting data and figures
Targets of the SARS-CoV-2 Neo Assay within the viral genome and detection channels
The positions of the different targets of the SARS-CoV-2 Neo Assay within the SARS-CoV-2 genome are displayed. Colors indicate the different dyes used for detection. Two targets are detected per channel of the qRT-PCR instrument.