QIAseq Targeted DNA Extended Panels
Digital DNA sequencing to confidently detect low-frequency variants
Digital DNA sequencing to confidently detect low-frequency variants
Cat. No. / ID: 333545
The QIAseq Targeted DNA Panels have been developed as a complete Sample to Insight solution to enable digital DNA sequencing by utilizing molecular barcodes. Digital DNA sequencing is a unique approach to detect low-frequency variants with high confidence by overcoming the issues of PCR duplicates, false positives and library bias.Each panel is a one-box, NGS platform-agnostic solution that contains all the necessary components to construct libraries from enriched genomic targets. Primer design is based on QIAseq Enrichment Technology, in which each genomic target is enriched by one target-specific primer and one universal primer – a strategy that removes conventional two target-specific primer design restriction and reduces the amount of required primers. All primers required for a panel are pooled into an individual primer pool to reduce panel handling and the number of pools required for enrichment and library construction. Platform-specific indexes, which are contained in a separate box, allow the multiplexing of up to 384 samples per sequencing run. The unique buffer and enzyme system used in the QIAseq targeted DNA panels has been optimized to achieve high coverage of GC-rich genomic regions.
Extended panels allow you to extend the content of any of the cataloged panels by adding up to 100 new primers. The new primers will be added to the content of the cataloged panel.
Need a quote for your research project or would you like to discuss your project with our specialist team? Contact Us
PCR duplicates are a major issue in targeted DNA sequencing, since, through PCR amplification, they turn unique DNA molecules into identical DNA molecules that cannot be distinguished from each other. In addition, errors from PCR amplification and sequencing process may also be present in final reads that lead to false positive variants in sequencing results. This, in turn, results in the inability to confidently call DNA variants present at low frequencies in the starting DNA material. To overcome the issue of PCR duplicates and amplification artifacts, the QIAseq Targeted DNA Panels use digital sequencing by incorporating molecular barcodes into the starting DNA material before any amplification takes place, thereby preserving the uniqueness of the starting DNA molecules and overcoming the issues of PCR duplicates, false positives and library bias.
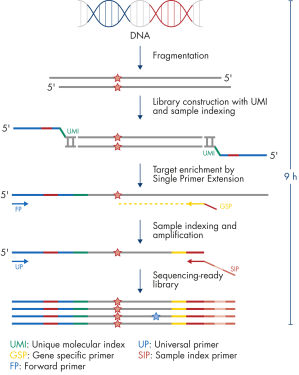
The entire workflow of the QIAseq Targeted DNA Panels to go from extracted DNA to sequencing-ready libraries can be completed in 9 hours. Extracted DNA is fragmented, genomic targets are molecularly barcoded and enriched, and libraries are constructed. Sequencing files can be fed into the QIAseq pipeline, a cloud-based data analysis pipeline, which will filter, map and align reads, as well as count unique molecular barcodes associated with targeted genomic regions, and call variants with a barcode-aware algorithm. This data can then be fed into IVA or QCI for interpretation.
The QIAseq Targeted DNA Extended Panels can be used to call a variety of DNA variants from a wide range of sample types for numerous applications.
DNA variants:
Sample types:
Applications:
Isolated DNA, as low as 20 ng, is enzymatically fragmented to generate small pieces of dsDNA. This is followed by the library construction step, in which IL-N7 adapters, molecular barcodes, and sample indexes are incorporated into DNA fragments generated in the previous step. Library fragments now serve as templates for target enrichment using single primer extension. In this step, targets are enriched using a single gene-specific primer and a universal forward primer. The final step is library amplification and sample indexing (for dual indexing) using the IL-S5 sample index primer and a universal primer.