✓ 연중무휴 하루 24시간 자동 온라인 주문 처리
✓ 풍부한 지식과 전문성을 갖춘 제품 및 기술 지원
✓ 신속하고 안정적인 (재)주문
MinElute Gel Extraction Kit (50)
Cat. No. / ID: 28604
✓ 연중무휴 하루 24시간 자동 온라인 주문 처리
✓ 풍부한 지식과 전문성을 갖춘 제품 및 기술 지원
✓ 신속하고 안정적인 (재)주문
특징
- 매우 적은 용출량
- 신속한 절차 및 간편한 취급 방법
- 재현성이 뛰어난 높은 회수율
- 편리한 샘플 분석을 위한 겔 로딩 염료
제품 세부 정보
MinElute Gel Extraction Kit는 최대 400mg의 겔 절편에서 70bp~4kb의 DNA 절편을 실리카 막 기반으로 정제할 수 있는 스핀 컬럼, 완충액, collection 튜브를 제공합니다. 스핀 컬럼은 매우 적은 용량(최소 10µl)으로 용출할 수 있도록 설계되어 고농축 DNA를 높은 수율로 추출할 수 있습니다. 포함된 pH indicator 염료를 통해 스핀 컬럼에 결합하는 DNA에 대한 최적의 pH를 쉽게 파악할 수 있습니다. MinElute system으로 정제된 DNA 절편은 염기서열 분석, 마이크로어레이 분석, 결찰(ligation) 및 형질전환, 제한효소 처리(restriction digestion), 라벨링, 미세 주입, PCR 및 체외 전사를 포함한 모든 응용 분야에서 바로 사용할 수 있습니다. MinElute Gel Extraction Kit는 QIAcube Connect에서 자동화할 수 있습니다.
최적의 결과를 얻으려면 이 제품을 QIAvac 24 Plus와 함께 사용하는 것이 좋습니다.
성능
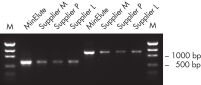
MinElute Gel Extraction Kit는 DNA 샘플에서 뉴클레오타이드, 효소, 염, 아가로스, 브로민화 에티듐(ethidium bromide) 및 기타 불순물을 제거하여 다양한 다운스트림 응용 분야에 적합한 고농축 DNA를 제공합니다(그림 " 고농축 DNA" 참조).
MinElute Gel Extraction Kit는 겔 추출을 위한 스핀 컬럼을 제공합니다. 마이크로 원심분리기 또는 진공 매니폴드를 사용하여 70bp~4kb의 고농도 DNA를 빠르게 정제합니다. (4kb~10kb의 DNA 절편은 QIAquick Gel Extraction Kit를 사용하여 정제해야 하며, 70bp보다 작거나 10kb보다 큰 DNA 절편은 QIAEX II Gel Extraction System으로 추출해야 합니다).
그림 참조
원리
MinElute Kit에는 염도가 높은 완충액에서 DNA를 결합하고 저염 완충액 또는 물로 용출하기 위한 실리카 막 어셈블리가 포함되어 있습니다. 실리카 막 기술은 분산 수지 및 슬러리(slurry)와 관련된 문제와 불편을 해소합니다. 특수 결합 완충액은 각 응용 분야에 최적화되어 있으며 특정 크기 범위 내에서 DNA 분자를 선택적으로 흡착하도록 합니다.
겔 로딩 염료
더 빠르고 편리한 샘플 처리 및 분석을 위해 겔 로딩 염료가 제공됩니다. GelPilot 로딩 염료는 아가로스 겔 실행 시간을 최적화하고 작은 DNA 절편이 너무 멀리 이동하는 것을 방지하기 위해 세 가지 추적 염료(크실렌 시아놀, 브로모페놀 블루, 오렌지 G)를 함유하고 있습니다(그림 " GelPilot 로딩 염료" 참조).
그림 참조
절차
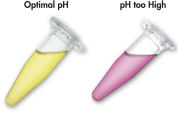
MinElute system은 간단한 결합-세척-용출 절차를 사용합니다(순서도 " MinElute 절차" 참조). 겔 절편을 pH indicator 염료가 포함된 완충액에 용해하여 DNA 결합을 위한 최적의 pH를 쉽게 확인할 수 있으며(그림 " pH indicator 염료" 참조), 혼합물을 MinElute pin column에 적용합니다. 핵산은 완충액의 높은 염도 조건에서 실리카겔 막에 흡착됩니다. 불순물을 씻어내고 제공된 소량의 저염 완충액 또는 물로 순수한 DNA를 용출하여 후속 응용 분야에 바로 사용할 수 있습니다.
취급
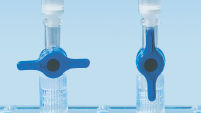
MinElute 스핀 컬럼은 두 가지 편리한 취급 옵션을 제공하도록 고안되었습니다. 스핀 컬럼은 기존의 탁상용 마이크로 원심분리기 또는 루어 커넥터가 있는 모든 진공 매니폴드(예: QIAvac Luer Adapters가 있는 QIAvac 24 Plus)에 장착할 수 있습니다. MinElute Gel Extraction Kit는 다른 QIAGEN 스핀 컬럼 기반 키트와 더불어 QIAcube Connect에서 완전히 자동화할 수 있어 생산성을 높이고 결과를 표준화할 수 있습니다 (그림 "스핀 컬럼 취급 옵션 A, B, C, D 및 QIAcube Connect" 참조).
그림 참조
응용 분야
MinElute 또는 QIAquick System으로 정제된 DNA 절편은 다음을 포함한 모든 응용 분야에서 바로 사용할 수 있습니다.
- 차세대 염기서열 분석을 포함한 염기서열 분석
- 마이크로어레이 분석
- 결찰(ligation) 및 형질전환
- 제한효소 처리(Restriction digestion)
- 라벨링
지원되는 데이터 및 수치
스핀 컬럼 취급 옵션 — C.

Specifications
| Features | Specifications |
|---|---|
| Binding capacity | 5µg |
| Elution volume | 10µL |
| Fragment size | 70bp~4kb |
| Sample type: applications | DNA: PCR 반응 |
| Technology | 겔 추출 |
| Recovery: oligonucleotides dsDNA | 회수: dsDNA 절편 |
| Format | 튜브 |
| Processing | 수동 |