Products
Features
- Compliant with EU IVD Directive 98/79/EC
- Reliable quantification of allele representation and methylation status
- Sequence information enables discovery of rare mutations
- More than one assay can be completed in a single run
- 1–24 samples can be analyzed in as little as 15 minutes
Product Details
The PyroMark Q24 MDx uses proven Pyrosequencing technology for real-time, sequence-based detection and quantification for in vitro diagnostic use in Europe. This innovative platform is highly suited for genotyping mutations, evaluating disease-related DNA methylation patterns, validating biomarkers, and other diagnostic-related assays.
Performance
Pyrosequencing technology enables accurate and sensitive quantification of genetic and epigenetic DNA variations by providing highly reliable sequence data. It allows the identification of novel mutations, as well as detection of aberrant DNA methylation patterns present at low levels.
The therascreen KRAS Pyro Kit is a good example of the informative analysis provided by the therascreen Pyro Kits and the PyroMark Q24 MDx instrument. The KRAS gene is mutated in approximately 35% of metastatic colorectal cancer (CRC) patients. Studies have shown that KRAS mutation testing can better define which CRC patients will benefit from treatment with epidermal growth factor receptor (EGFR) inhibiting monoclonal antibodies, such as panitumumab and cetuximab.
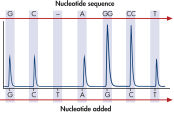
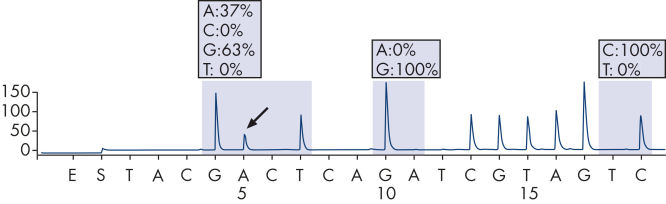
The therascreen KRAS Pyro Kit consists of 2 assays: one for detecting mutations in codons 12 and 13 and the other for detecting mutations in codon 61. The two regions are amplified separately by PCR, using optimized PCR reagents and primers included in the kit, and then sequenced through the defined region (see figure " Normal genotype in codons 12 and 13"). Sequences surrounding the defined positions serve as normalization and reference peaks for quantification and quality assessment of the analysis. Pyrosequencing technology on the PyroMark Q24 MDx enables identification of specific mutations (see figure " GGT to GAT mutation in base 2 of codon 12"), including less frequent mutations (see figures " GGT to AGT mutation in base 1 of codon 12" and " Reanalysis of the data in figure 'GGT to AGT mutation in base 1 of codon 12'"), as well as discovery of new mutations.
See figures
Principle
Pyrosequencing technology, which is based on the principle of sequencing by synthesis, provides quantitative data in sequence context within minutes. PyroMark Q24 MDx is a fully integrated system that provides real-time sequence information and is highly suited for genetic and epigenetic analysis. The system includes the PyroMark Q24 MDx Instrument, PyroMark Q24 MDx Vacuum Workstation, PyroMark Q24 MDx Software 2.0, PyroMark Gold Q24 Reagents, PyroMark Control Oligo, and PyroMark Q24 Validation Oligo. Sample preparation solutions are also available to enable preparation of single-stranded DNA using the PyroMark Q24 MDx Vacuum Workstation.
Steps of the Pyrosequencing reaction:
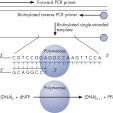
Step 1: A DNA segment is amplified, and the strand to serve as the Pyrosequencing template is biotinylated. After denaturation, the biotinylated single-stranded PCR amplicon is isolated and allowed to hybridize with a sequencing primer. The hybridized primer and single-stranded template are incubated with the enzymes DNA polymerase, ATP sulfurylase, luciferase, and apyrase, as well as the substrates adenosine 5' phosphosulfate (APS) and luciferin (see figure " Principle of Pyrosequencing — step 1").
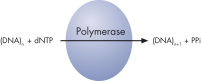
Step 2: The first deoxribonucleotide triphosphate (dNTP) is added to the reaction. DNA polymerase catalyzes the addition of the dNTP to the squencing primer, if it is complementary to the base in the template strand. Each incorporation event is accompanied by release of pyrophosphate (PPi), in a quantity equimolar to the amount of incorporated nucleotide (see figure " Principle of Pyrosequencing — step 2").
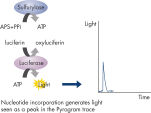
Step 3: ATP sulfurylase converts PPi to ATP in the presence of adenosine 5' phosphosulfate (APS). This ATP drives the luciferase-mediated conversion of luciferin to oxyluciferin that generates visible light in amounts that are proportional to the amount of ATP. The light produced in the luciferase-catalyzed reaction is detected by CCD sensors and seen as a peak in the raw data output (Pyrogram). The height of each peak (light signal) is proportional to the number of nucleotides incorporated (see figure " Principle of Pyrosequencing — step 3").
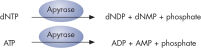
Step 4: Apyrase, a nucleotide-degrading enzyme, continuously degrades unincorporated nucleotides and ATP. When degradation is complete, another nucleotide is added (see figure " Principle of Pyrosequencing — step 4").
Step 5: Addition of dNTPs is performed sequentially. It should be noted that deoxyadenosine alfa-thio triphosphate (dATPαS) is used as a substitute for the natural deoxyadenosine triphosphate (dATP), since it is efficiently used by the DNA polymerase, but not recognized by the luciferase. As the process continues, the complementary DNA strand is elongated, and the nucleotide sequence is determined from the signal peaks in the Pyrogram trace (see figure " Principle of Pyrosequencing — step 5").
Dedicated IVD-validated assays
QIAGEN offers an expanding suite of IVD-validated assays to be used with the PyroMark Q24 MDx. Currently, these kits include important cancer-related mutations:
| Product | For quantitative measurement of mutations in |
|---|---|
| therascreen KRAS Pyro Kit | codons 12, 13, and 61 of the human KRAS gene |
| therascreen BRAF Pyro Kit | codons 600 and 464-469 of the human BRAF gene |
| therascreen EGFR Pyro Kit | codons 719, 768, 790, 858, and 861 and exon 19 of the human EGFR gene |
| therascreen NRAS Pyro Kit | codons 12, 13, and 61 of the human NRAS gene |
See figures
Procedure
From PCR product to single-stranded template ready for sequencing — up to 24 samples can be prepared in parallel using the PyroMark Q24 MDx Vacuum Workstation, in less than 15 minutes. The workstation ensures easy handling, and the actual hands-on time is less than 5 minutes.
Prior to Pyrosequencing, a biotinylated PCR product is generated. This biotinylated PCR product is bound to Streptavidin-coated Sepharose beads, and the beads are captured with the Vacuum Tool on the Vacuum Workstation, where they are thoroughly washed and subsequently denatured, generating single-stranded DNA suitable for Pyrosequencing. This template DNA is released into the Pyrosequencing reaction plate containing the sequencing primer, and after primer annealing, the plate is placed into the PyroMark instrument. PyroMark Gold reagents contain the enzymes, nucleotides, and substrate for the Pyrosequencing reaction; these are pipetted into the dispensing cartridge, according to the volumes provided by the software, and are also placed into the instrument for the Pyrosequencing run.
Applications
The PyroMark Q24 MDx is suitable for in vitro diagnostic applications in Europe.
Genetic analysis comprises multiple applications to analyze differences in genomic DNA, including mutation detection and SNP typing. PyroMark Q24 MDx facilitates accurate and highly sensitive mutational analysis of any gene of interest and enables quantification of allele representation in mixed cell populations. The system can be used for self-validated assays for oncology studies and for analysis of epigenetic markers in methylation studies, or it can be used with dedicated IVD-validated assays available from QIAGEN
Software
PyroMark Q24 MDx Software, installed on a PC, enables comprehensive analysis of your results. The software contains two analysis modes: CpG and AQ (allele quantification). Both modes can be used to analyze samples on the same plate, enabling different types of samples to be run at the same time. The AQ mode can be used for analyzing single and multivariable positions, as well as di-, tri- , and tetra- allelic mutations. The CpG mode enables analysis of multiple consecutive CpG sites and provides a built-in control for the bisulfite treatment.
Supporting data and figures
GGT to GAT mutation in base 2 of codon 12.
Specifications
| Features | Specifications |
|---|---|
| Connections | One USB port (2.0) |
| Chemical resistance | pH 4 to pH 9, common detergents, 0.5 M sodium hydroxide, ethanol |
| Applications | Methylation analysis, allele quantification, genotyping, sequence analysis |
| Humidity | Relative humidity of 20–90% (noncondensing) |
| CE/FDA/IVD compatible | In Europe |
| Instrument dimensions | 420 x 390 x 525 mm (16.5 x 15.4 x 20.7 in.) |
| Kits designed for this instrument | IVD-labeled therascreen Kits |
| Operating temperature | 15–32°C (59–90°F) |
| Overvoltage category | II |
| Place of operation | For indoor use only |
| Pollution level | 2 |
| Power | 100–240 V AC, 47–63 Hz, 1.1–0.45 A (grounded). From external power supply to instrument: 12 VDC and 24 VDC nominal |
| Process temperature | 28°C (82.4°F) ± 1°C |
| Process time | Depends on the number of nucleotide dispensations (20 dispensations take 24 minutes) |
| Samples per run (throughput) | 1–24 |
| Software | PyroMark Q24 MDx Software 2.0 |
| Technology | Pyrosequencing |
| Weight | 27.5 kg (60.6 lb) |
| Altitude | Up to 2000 m (6500 ft) |