Total DNA샘플에 포함된 human mitochondrial DNA로 부터 신뢰성있는 whole genome amplification이 가능합니다
✓ 연중무휴 하루 24시간 자동 온라인 주문 처리
✓ 풍부한 지식과 전문성을 갖춘 제품 및 기술 지원
✓ 신속하고 안정적인 (재)주문
REPLI-g Mitochondrial DNA Kit (25)
Cat. No. / ID: 151023
✓ 연중무휴 하루 24시간 자동 온라인 주문 처리
✓ 풍부한 지식과 전문성을 갖춘 제품 및 기술 지원
✓ 신속하고 안정적인 (재)주문
특징
- 정확하고 재현성있는 결과 — even from small total DNA sample
- Versatility — 머리카락을 포함하여 다양한 sample material에서 부터 DNA의 분리 정제 가능
- 빠르고 손쉬운 프로토콜 — kit 안에 모든 시약이 포함
제품 세부 정보
The REPLI-g Mitochondrial DNA Kit enables selective amplification of mitochondrial DNA from total DNA samples, without the need for prior mitochondrial DNA isolation. This kit provides DNA polymerase, buffers, and reagents for specific and uniform whole genome amplification from small samples of human mitochondrial DNA in total DNA samples using multiple displacement amplification (MDA). The simple procedure enriches for mitochondrial DNA with minimal contamination from nuclear DNA, thus avoiding the need for time-consuming isolation of mitochondrial DNA and increasing the sensitivity of downstream assays.
성능
그림 참조
원리
One limitation to the use of mitochondrial DNA has been the need to isolate it from nuclear DNA, particularly in cases where the sensitivity of mitochondrial DNA marker assays needs to be increased. The isolation procedure involves many time-consuming steps and leads to substantial losses of mitochondrial DNA. The REPLI-g Mitochondrial DNA Kit overcomes this limitation by enriching for mitochondrial DNA sequences in total DNA samples.
REPLI-g Mitochondrial DNA technology provides fast and highly uniform DNA amplification across the entire mitochondrial genome. The method is based on multiple displacement amplification (MDA) technology, which carries out isothermal genome amplification utilizing a uniquely processive DNA polymerase. Due to its high processivity and strand displacement activity, REPLI-g DNA Polymerase is capable of amplifying up to 100 kb without dissociating from the DNA template and minimizes unequal sequence and locus representation compared with PCR-based amplification methods.
절차
그림 참조
응용 분야
REPLI-g amplified mitochondrial DNA can be used in a variety of downstream applications, including:
- Genotyping (e.g., SNP, deletions, insertions)
- Endpoint-PCR, quantitative real-time PCR
- Sequence analysis
지원되는 데이터 및 수치
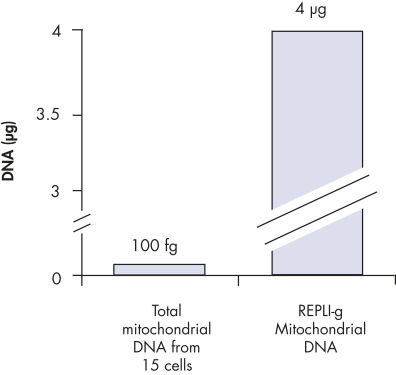
Enrichment of mitochondrial DNA.
Specifications
| Features | Specifications |
|---|---|
| Amplification | Whole genomic DNA |
| Samples per run (throughput) | Mid |
| Denaturation step | Heat |
| Maximum input volume | 10 µl template DNA |
| Minimal pipetting volume needed | 1 µl |
| Reaction volume | 50 µl |
| Reaction time | ~8 hours (overnight) |
| Quality assessment | No |
| Applications | Genotyping, sequencing, RFLP |
| Starting amount of DNA | ~10 ng purified total DNA |
| Starting material | Genomic human DNA |
| Technology | Multiple Displacement Amplification (MDA) |
| Yield | ~4 µg |