Products

RT² SYBR Green Fluor qPCR Mastermix (24)
Cat. No. / ID. / ID. 330513

RT² SYBR Green qPCR Mastermix (12)
Cat. No. / ID. / ID. 330502

RT² SYBR Green ROX qPCR Mastermix (24)
Cat. No. / ID. / ID. 330523

RT² SYBR Green qPCR Mastermix (2)
Cat. No. / ID. / ID. 330500

RT² SYBR Green ROX qPCR Mastermix (25 ml)
Cat. No. / ID. / ID. 330529

RT² SYBR Green Fluor qPCR Mastermix (12)
Cat. No. / ID. / ID. 330512

RT² SYBR Green qPCR Mastermix (24)
Cat. No. / ID. / ID. 330503

RT² SYBR Green ROX qPCR Mastermix (6)
Cat. No. / ID. / ID. 330524

RT² SYBR Green Fluor qPCR Mastermix (2)
Cat. No. / ID. / ID. 330510

RT² SYBR Green qPCR Mastermix (25 ml)
Cat. No. / ID. / ID. 330509

RT² SYBR Green ROX qPCR Mastermix (8)
Cat. No. / ID. / ID. 330521

RT² SYBR Green qPCR Mastermix (8)
Cat. No. / ID. / ID. 330501

RT² SYBR Green Fluor qPCR Mastermix (25 ml)
Cat. No. / ID. / ID. 330519

RT² SYBR Green Fluor qPCR Mastermix (6)
Cat. No. / ID. / ID. 330514

RT² SYBR Green ROX qPCR Mastermix (12)
Cat. No. / ID. / ID. 330522

RT² SYBR Green Fluor qPCR Mastermix (8)
Cat. No. / ID. / ID. 330511

RT² SYBR Green qPCR Mastermix (6)
Cat. No. / ID. / ID. 330504

RT² SYBR Green ROX qPCR Mastermix (2)
Cat. No. / ID. / ID. 330520
Features
- Designed exclusively for use with RT2 and EpiTect Methyl II qPCR Assays
- Delivers specific amplification with minimal primer-dimers
- Available with ROX, fluorescein, or without dye
- For use with qPCR instruments under standard cycling conditions
Product Details
Performance
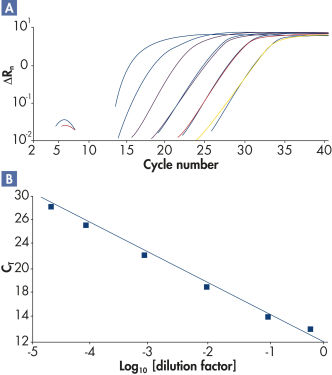
The high-performance real-time PCR enabled by RT² SYBR Green qPCR Mastermixes demonstrates high amplification efficiencies (see figure " High amplification efficiency over a wide dynamic range") and high levels of sensitivity and specificity (see figure " Tighter control of polymerase activity yields greater specificity").
See figures
Principle
RT2 SYBR Green qPCR Mastermixes contain real-time PCR buffer, a high-performance, HotStart DNA Taq polymerase, nucleotides, and SYBR Green dye. Some mastermixes contain either fluorescein or ROX dye for optimization of the instrument optics. The chemically-modified and tightly controlled HotStart enzyme uniquely provides accurate SYBR Green results by preventing the amplification of primer–dimers and other nonspecific products.
Mastermixes without ROX or fluorescein
RT2 SYBR Green qPCR Mastermix is highly suited for real-time PCR applications using SYBR Green-based detection on instrumentation not requiring a reference dye, including Bio-Rad models CFX96, CFX384; Bio-Rad/MJ Research Chromo4, DNA Engine Opticon, DNA Engine Opticon 2; Roche LightCycler 480 (96-well and 384-well); Eppendorf Mastercycler ep realplex without ROX filter set; and Cepheid SmartCycler.
Mastermixes with fluorescein
RT2 SYBR Green Fluor qPCR Mastermix is highly suited for real-time PCR applications using SYBR Green-based detection on instrumentation that uses fluorescein as a reference dye, including the Bio-Rad models iCycler, iQ5, MyiQ, and MyiQ2.
Mastermixes with ROX
RT2 SYBR Green ROX qPCR Mastermix is highly suited for real-time PCR applications using SYBR Green-based detection on instrumentation that uses ROX as a reference dye, including QIAGEN’s Rotor-Gene Q; Applied Biosystems models 5700, 7000, 7300, 7500 (Standard and Fast), 7700, 7900HT (Standard and Fast 96-well block, 384-well block), StepOnePlus, ViiA 7 (Standard and Fast 96-well block, 384-well block); Eppendorf Mastercycler ep realplex with or without ROX filter set; Stratagene models Mx3000P, Mx3005P, Mx4000; and Takara TP-800.
Applications
Supporting data and figures
High amplification efficiency over a wide dynamic range.