✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
QuantiTect Whole Transcriptome Kit (100)
Cat. No. / ID: 207045
✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
特徴
- 多数の定量PCR解析やアーカイブを実現する高いcDNA収量
- cDNAおよび転写物の全領域から均一に増幅
- 至適化された試薬で迅速かつ簡単な実験操作
製品詳細
QuantiTect Whole Transcriptome Kit では、限られた量のRNAから増幅および逆転写反応により高収量のcDNAのを得ることができ、リアルタイムPCRによる多数の遺伝子発現解析が可能になります。至適化済みのプロトコールにより迅速かつ簡単に操作できます。本キットは、全トランスクリプトーム増幅(whole transcriptome amplification;WTA)のための酵素および試薬のセットです。Phi29 ポリメラーゼテクノロジーの画期的な改良により、わずか1 ng のRNAから最高40 μg のcDNAが得られます。このポリメラーゼの画期的な高い伸長性により、均一に増幅されたターゲットを含むcDNAが合成され、リアルタイムPCRで信頼性のある遺伝子発現解析を実現します。
パフォーマンス
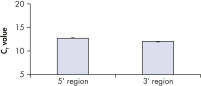
QuantiTect Whole Transcriptome Kit による全転写物の均一な増幅は、信頼できる遺伝子発現解析において必須条件です。全てのmRNA転写物が5'領域および3'領域の両方で均一に増幅されます(図 " 5’領域および3’領域を均一に増幅")。QuantiTect Whole Transcriptome Kit でWTA増幅したcDNA と、逆転写酵素で調製した増幅なしのcDNAを比較した結果を図 " 転写物プロファイルの保持"に示しています。これは、転写物プロファイルが保持されていることを証明しています。
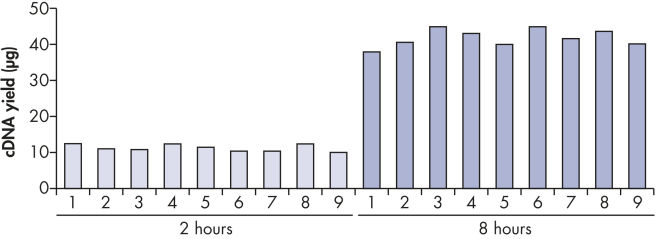
最高40 μg までの高収量cDNAが再現性良く得られます(図 " 一定したcDNA収量")。この特長によりリアルタイムPCR 解析を何回も行なうことが可能で、CT 値も一定しています(表および図 " 信頼できるリアルタイムPCR解析")。さらにcDNAはRNAよりも安定であるため、将来行なう研究実験用に、安全に保存することができます。
| スタートサンプル | 増幅時間 | 増幅産物の一般的な収量 | 解析可能なリアルタイムPCR解析の回数* |
|---|---|---|---|
| 10 ng RNA | 2 時間 | 最高 10 µg/反応 | 1000 |
| 10 ng RNA | 8 時間 | 最高 40 µg/反応 | 4000 |
図参照
原理
使用可能な生体サンプルが微量の場合、遺伝子発現プロファイル解析が制限されることがあります。QuantiTect Whole Transcriptome Kitを用いることで、微量かつ貴重なサンプルから膨大な数のリアルタイムPCR 解析が可能になります。QuantiTect Whole Transcriptome Kitは、わずか1 ng のRNA あるいは約50個の細胞に相当するRNAからの全トランスクリプトーム増幅用に至適化されています。RNAの品質および目的の転写物の量によっては、さらに少量のRNAでも使用できます。
QuantiTect Whole Transcriptome Kitには、RNAサンプル中の全転写物の増幅のための逆転写酵素、DNAポリメラーゼ、至適化済みバッファーと試薬が含まれています。本キットは、バイアスのない配列増幅のために、実証済みのREPLI-g テクノロジーを用いたcDNA合成を組み込んでいます。REPLI-g 増幅は、伸長性が高いDNAポリメラーゼを用いて等温でゲノム増幅ができるMultiple Displacement Amplification(MDA)テクノロジーを利用しています(図 " 全トランスクリプトーム増幅)。本テクノロジーは転写物の全領域(5'領域からでも)からバイアスのない均一な増幅を実現するので、リアルタイムPCRに最適な高収量のcDNAが得られます。
図参照
操作手順
図参照
アプリケーション
QuantiTect Whole Transcriptome Kit を用いて合成したcDNAはQuantiFastやRotor-Gene Kitsを用いたリアルタイムPCR解析に使用でき、将来行なう解析のために長期保存することもできます。このcDNAはマイクロアレイ解析には適切ではありません。少量あるいは貴重なサンプルからの全ゲノム増幅に、QIAGENはREPLI-g kitsを提供しています。配列によるバイアスは最小限で均一に増幅されるので、増幅したDNAはジェノタイピングやCGH(Comparative Genome Hybridization)のようなアプリケーションに最適です。
裏付けデータと数値
Reproducible cDNA yields.
Specifications
| Features | Specifications |
|---|---|
| Amplification | Whole total RNA |
| Starting material | Purified total RNA |
| Reaction time | 5 hours or 11 hours |
| Applications | Real-time PCR, gene expression analysis |
| Minimal pipetting volume needed | 1 µl |
| Samples per run (throughput) | Mid |
| Technology | Reverse transcription followed by ligation and multiple displacement amplification (MDA) |
| Yield | 10 µg or 40 µg cDNA |
| Maximum input volume | 5 µl total RNA |
| Starting amount of total RNA | >10 ng purified total RNA |
| Reaction volume | 50 µl |