✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
QuantiNova Reverse Transcription Kit (50)
Cat. No. / ID: 205411
For 50 x 20 µl reactions: 100 µl 8x gDNA Removal Mix, 50 µl Reverse Transcription Enzyme, 200 µl Reverse Transcription Mix (containing RT primers), 100 µl Internal Control RNA, 1.9 ml RNase-Free Water
ログイン アカウントの価格を確認するには
KitAssay
QuantiNova Reverse Transcription Kit
QuantiNova IC Assay
Reactions
50
200
QuantiNova Reverse Transcription Kitは分子生物学的アプリケーション用であり、疾病の診断、予防、あるいは治療に使用することはできません。
✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
特徴
- gDNA除去によるアーティファクトな結果の削減
- 付属のインターナルコントロールを使用し、確実なcDNA合成のモニタリング
- 逆転写酵素の高いアフィニティーにより幅広いリニアレンジでcDNA合成が可能
製品詳細
ユニークなQuantiNova Reverse Transcription Kit はgDNA除去及び付属のインターナルコントロールを用いることで、迅速かつ簡便なcDNA合成のプロトコールを提供します。RNAサンプル中に混入しているゲノムDNAはgDNA Removal Mixによって除去され、かつ付属のInternal Control RNAによって、確実な逆転写と増幅を検証するために使用できます。QuantiNova Reverse Transcription Kit は迅速かつ効果的なcDNA合成を提供します。合成されたcDNAはリアルタイムPCRでの使用に至適化され、mRNA転写物の全ての領域を確実に定量できます。
パフォーマンス
Contaminating genomic DNA in RNA samples is effectively and rapidly removed with the unique gDNA Removal Mix. Elimination of genomic DNA is crucial for accurate gene expression results and when design of RNA-specific primers or probes is not always possible, for example, when analyzing single-exon genes. With gDNA Removal Buffer, time is saved and costs are reduced, since a separate DNase digestion during or after purification of RNA samples is not required.
The newly developed internal control is a defined transcript (RNA molecule) that can simply be added to your samples and transcribed into cDNA. It is intended to report instrument or chemistry failures, errors in assay setup and the presence of inhibitors. The expected range of Cq (CT) values can be used to test successful reverse transcription/amplification and the reproducibility of your results.
The high RNA affinity of the QuantiNova Reverse Transcriptase enables high yields of cDNA from any RNA template. Difficult templates, such as those with high GC-content or complex secondary structure, are also successfully reverse transcribed. The Reverse Transcription Mix contains a specially optimized mix of oligo-dT and random primers that enable cDNA synthesis from all regions of RNA transcripts, including 5' regions. The kit provides high yields of cDNA template for real-time PCR analysis, regardless of where the amplified target region is located on the transcript, and provides greater sensitivity in the detection of low-abundance genes.
The QuantiNova Reverse Transcription Kit offers accurate results over a wide range of input amounts, including up to 5 µg RNA in the standard protocol, which can be doubled to accommodate exceptionally large amounts of RNA.
The newly developed internal control is a defined transcript (RNA molecule) that can simply be added to your samples and transcribed into cDNA. It is intended to report instrument or chemistry failures, errors in assay setup and the presence of inhibitors. The expected range of Cq (CT) values can be used to test successful reverse transcription/amplification and the reproducibility of your results.
The high RNA affinity of the QuantiNova Reverse Transcriptase enables high yields of cDNA from any RNA template. Difficult templates, such as those with high GC-content or complex secondary structure, are also successfully reverse transcribed. The Reverse Transcription Mix contains a specially optimized mix of oligo-dT and random primers that enable cDNA synthesis from all regions of RNA transcripts, including 5' regions. The kit provides high yields of cDNA template for real-time PCR analysis, regardless of where the amplified target region is located on the transcript, and provides greater sensitivity in the detection of low-abundance genes.
The QuantiNova Reverse Transcription Kit offers accurate results over a wide range of input amounts, including up to 5 µg RNA in the standard protocol, which can be doubled to accommodate exceptionally large amounts of RNA.
原理
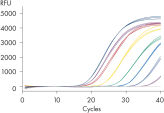
The QuantiNova Reverse Transcriptase has a high affinity for RNA and is capable of cDNA synthesis from a wide range of RNA amounts (10 pg – 5 μg) (see figure “ Precise quantification from 5 µg – 10 pg RNA). The kit provides high yields of cDNA template for real-time PCR analysis, regardless of where the amplified target region is located on the transcript. Even difficult templates, such as those with high GC-content or complex secondary structure, are successfully reverse transcribed. The QuantiNova RT Buffer has also been optimized to be compatible with real-time PCR buffers.
To obtain accurate results in real-time RT-PCR gene expression assays, it is important that only cDNA is amplified and detected. Interference by genomic DNA can effectively and rapidly be removed using the gDNA Removal Mix (see figure “ Efficient removal of contaminating gDNA ensures precise quantification of transcripts). Time is saved and costs are reduced, since a separate DNase digestion during or after purification of RNA samples is not required. Also, designing RNA-specific primers or probes is not necessary.
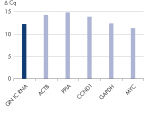
Detecting variations in cDNA synthesis allows you to check the reproducibility of your results. The newly developed internal control is a defined transcript (RNA molecule) that can be optionally added to your samples and transcribed into cDNA. It is intended to report instrument or chemistry failures, errors in assay setup and the presence of inhibitors. Because the internal control behaves comparable to real transcripts, it can be used to confirm successful reverse transcription and amplification in subsequent qPCR (see figure “ Internal control reliably detects presence of inhibitors).
To obtain accurate results in real-time RT-PCR gene expression assays, it is important that only cDNA is amplified and detected. Interference by genomic DNA can effectively and rapidly be removed using the gDNA Removal Mix (see figure “ Efficient removal of contaminating gDNA ensures precise quantification of transcripts). Time is saved and costs are reduced, since a separate DNase digestion during or after purification of RNA samples is not required. Also, designing RNA-specific primers or probes is not necessary.
Detecting variations in cDNA synthesis allows you to check the reproducibility of your results. The newly developed internal control is a defined transcript (RNA molecule) that can be optionally added to your samples and transcribed into cDNA. It is intended to report instrument or chemistry failures, errors in assay setup and the presence of inhibitors. Because the internal control behaves comparable to real transcripts, it can be used to confirm successful reverse transcription and amplification in subsequent qPCR (see figure “ Internal control reliably detects presence of inhibitors).
| Component | Benefits |
|---|---|
| gDNA Removal Mix | Detection of RNA only in real-time RT-PCR |
| Internal Control RNA | Verification of successful RT-PCR performance |
| QuantiNova Reverse Transcriptase | Use of a wide range of RNA amounts (10 pg – 5 μg RNA) High sensitivity |
| QuantiNova RT buffer system | Read-through of difficult templates |
| RT Primer Mix | cDNA synthesis from all regions of transcripts, even from 5' regions |
図参照
操作手順
Genomic DNA removal and cDNA synthesis take only 20 minutes with the QuantiNova Reverse Transcription Kit. The kit includes everything you need for fast cDNA synthesis.
Purified RNA is briefly incubated in gDNA Removal Mix to effectively remove contaminating genomic DNA. In contrast to other methods, the RNA sample is then used directly for reverse transcription in combination with a master mix prepared from the Reverse Transcription Mix plus Reverse Transcription Enzyme. With the QuantiNova Reverse Transcriptase, RNA can be transcribed at low temperatures, even through complex 2° degree structure. This ensures that the RNA will stay intact as the reaction takes place at 42°C and is then inactivated at 85°C. Additional steps for RNA denaturation or RNase H digestion are not necessary.
Purified RNA is briefly incubated in gDNA Removal Mix to effectively remove contaminating genomic DNA. In contrast to other methods, the RNA sample is then used directly for reverse transcription in combination with a master mix prepared from the Reverse Transcription Mix plus Reverse Transcription Enzyme. With the QuantiNova Reverse Transcriptase, RNA can be transcribed at low temperatures, even through complex 2° degree structure. This ensures that the RNA will stay intact as the reaction takes place at 42°C and is then inactivated at 85°C. Additional steps for RNA denaturation or RNase H digestion are not necessary.
アプリケーション
The QuantiNova Reverse Transcription Kit allows highly efficient and sensitive real-time RT-PCR for all types of starting materials, including laser-microdissected samples and tissue biopsies.
裏付けデータと数値
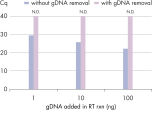
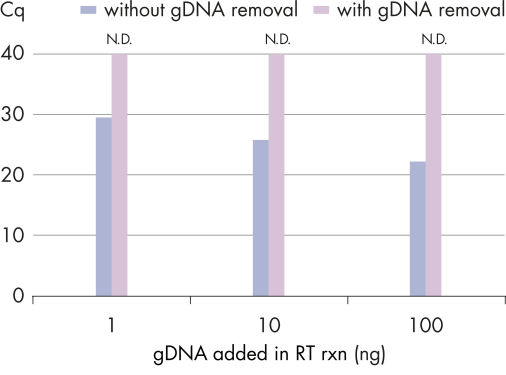
Efficient removal of contaminating gDNA ensures precise quantification of transcripts.
Real-time two-step RT-PCR analysis of ACTB, with and without gDNA removal step. Instead of using RNA, genomic DNA was purified from human whole blood and used as template. Different amounts of gDNA (100 ng, 10 ng, 1 ng) were spiked into the cDNA synthesis reactions using the QuantiNova Reverse Transcription Kit. Real-time PCR was performed in triplicate on an Applied Biosystems 7500 Fast Real-Time PCR System using a 1:10 cDNA dilution and the QuantiNova Probe PCR Kit. Without gDNA removal, genomic DNA contamination resulted in a strong signal. This signal was successfully eliminated by the integrated gDNA removal step (N.D. = not detected).
リソース
パンフレット (6)
セレクションガイド (1)
キットハンドブック (3)
クイックスタートプロトコール (2)
MSDS (1)
Safety Data Sheets (1)
Certificates of Analysis (1)
Brochures & Guides (6)
Kit Handbooks (3)
Selection Guides (1)
Quick-Start Protocols (2)