RT2 SYBR® Green FAST Mastermixes
リアルタイムPCRアプリケーション用
リアルタイムPCRアプリケーション用

Cat. No. / ID: 330620
RT² SYBR Green FAST Mastermixesには、RT2 qPCR Primer AssayおよびRT2 Profiler PCR Arrayを用いたSYBR GreenベースのリアルタイムPCRに必要な至適化済み試薬とバッファーがすべて入っていて、ほとんどのリアルタイムPCR装置で使用できます。
RT² SYBR Green FAST Mastermixesを用いて行なった高性能リアルタイムPCRは、高い増幅効率(図“ 幅広いダイナミックレンジで高い増幅効率”)および高レベルの感度と特異性(図“ ポリメラーゼ活性の厳密な制御により高い特異性を実現”)を実証しています。
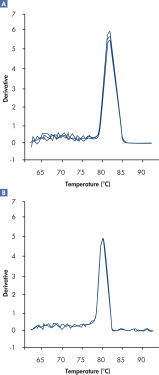
さらにRT2 SYBR Green FAST Mastermixesを用いて得られた解離曲線は、通常のサイクリング条件で得られる解離曲線に比べてバックグランドが低く、より明確な結果が得られます(図“ RT2 SYBR Green Fast Mastermixを用いて明確な解離曲線を実現”)。
RT2 SYBR Green FAST Mastermixesには、リアルタイムPCRバッファー、高性能HotStart DNA Taq polymerase、ヌクレオチド、SYBR Green色素が入っています。装置の光学系を最適化するために、フルオレセインまたはROX色素のいずれかが入ったマスターミックスもあります。化学修飾され厳密にコントロールされたHotStart酵素を用いることにより、プライマーダイマーあるいは非特異的な産物の増幅を回避でき、正確なSYBR Green結果が得られます。RT2 SYBR Green FAST Mastermixesは、10秒間の変性ステップ、30秒間のアニーリング/エクステンションステップをもつPCRプロトコール用に至適化され、特異性や感度の低下はありません。
RT2 SYBR Green FAST Mastermixは reference dyeが不要な装置でのSYBR Greenベースの検出用リアルタイムPCRアプリケーションに最適です。 対象装置: Bio-Rad models CFX96、CFX384; Bio-Rad/MJ Research Chromo4、DNA Engine Opticon、DNA Engine Opticon 2; Roche LightCycler 480 (96-well and 384-well); Eppendorf Mastercycler ep realplex without ROX filter set; Cepheid SmartCycler。
RT2 SYBR Green Fluor FAST Mastermixは、 reference dyeとしてフルオレセインを使用する装置でのSYBR Greenベースの検出用リアルタイムPCRアプリケーションに最適です。 対象装置:Bio-Rad models iCycler、iQ5、MyiQ、MyiQ2。
RT2 SYBR Green ROX FAST Mastermixは、 reference dyeとしてROXを使用する装置でのSYBR Greenベースの検出用アルタイムPCRアプリケーションに最適です。 対象装置:QIAGENの Rotor-Gene Q; Applied Biosystems models 5700, 7000, 7300, 7500 (Standard and Fast), 7700, 7900HT (Standard and Fast 96-well block, 384-well block), StepOnePlus, ViiA 7 (Standard and Fast 96-well block, 384-well block); Eppendorf Mastercycler ep realplex with or without ROX filter set; Stratagene models Mx3000P, Mx3005P, Mx4000;Takara TP-800。
A positive PCR control was amplified on the ABI 7500 qRT-PCR instrument using [A] regular cycling and RT² SYBR Green Mastermix or [B] fast cycling with RT² SYBR Green FAST Mastermix. The dissociation curve obtained with RT² SYBR Green FAST Mastermix has lower background and a sharper shape.