QuantiTect Probe PCR Kit (200)
Cat. No. / ID: 204343
特徴
- 低コピーターゲットを高感度で検出
- 配列特異的プローブまたはSYBR Greenを使用するqPCR
- テンプレートの複数のログにわたる正確な定量
- 反応条件やサイクリング条件の最適化が不要
- 同じチューブで最大5標的を検出
製品詳細
QuantiTect PCR Kits(qPCRキット)は、real-time PCRと配列特異的プローブまたはSYBR Green I検出を使用する2ステップRT-PCRによって、gDNAおよびcDNA標的の高感度定量を可能にします。また、real-time PCRキットでは、1本のチューブで、マルチプレックス、real-time PCR、または2ステップRT-PCRによって、最大5つのgDNAまたはcDNA標的を確実に定量できます。すぐに使えるマスターミックスにホットスタートと独自のPCRバッファーシステムを組み合わせることで、最適化の必要なく、どのリアルタイムサイクラーでも高感度のqPCRが保証されます。dNTPミックスにはdUTPが含まれており、オプションでUNGによる処理が可能です。QuantiTect PCR Kitのマスターミックスは2–8°Cで保存でき、便利です。
配列特異的プローブを使用するマルチプレックスPCRには、蛍光正規化にROX色素を必要とするサイクラー用のQuantiTect Multiplex PCR Kitと、その他すべてのサイクラー用のQuantiTect Multiplex PCR NoROX Kitという2種類のキット形式があります。
パフォーマンス
QuantiTect定量PCRキットに含まれるHotStarTaq DNA Polymeraseは、他のポリメラーゼに比べ、最も厳密なホットスタートを提供することによって、PCR反応の特異性を高めます。
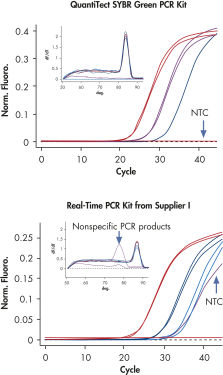
QuantiTect SYBR Green PCR Kitsは、広い直線範囲で特異的定量を可能にします(図「 広い直線範囲での特異的定量」参照)。QuantiTect Reverse Transcription KitおよびQuantiTect Primer Assaysと組み合わせて使用すると、QuantiTect SYBR Green PCR Kitsは高感度で信頼性の高い結果を提供します(図「 特異的で高感度な定量」参照)。
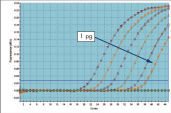
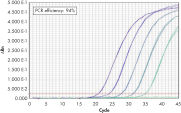
QuantiTect Probe PCR Kitsは、QuantiTect Reverse Transcription Kitと組み合わせると、高感度で信頼性の高い結果を提供します(図「 高い感度と効率、広いダイナミックレンジ」参照)。独自のPCRバッファー組成によって、QuantiTect Probe PCR Kitsは低コピーDNA標的の高感度定量だけでなく、広い直線範囲での正確な定量を実現します(図「 real-time PCRにおける広いダイナミックレンジ」参照)。
QuantiTect Multiplex PCR Master Mixは、マルチプレックス反応のPCR産物が、対応するシングル増幅反応のPCR産物と同じ効率と感度で増幅されることを保証します(表“トリプレックスPCRおよびシングルプレックスPCRと同等の閾値サイクル(CΤ)”と図“ 4-プレックスおよびシングルプレックスPCRと同等の結果”参照)。
| t(8;14)転座配列の検出(20コピー) | GAPDH cDNA配列の検出(106コピー) | NFKB cDNA配列の検出(コピー数については最初の列を参照) | |
|---|---|---|---|
| 105コピーのNFKBを用いるトリプレックスPCR | 34.31 | 20.37 | 21.92 |
| 対応するシングルプレックスPCR | 34.07 | 20.54 | 21.83 |
| 104コピーのNFKBを用いるトリプレックスPCR | 34.61 | 20.62 | 25.03 |
| 対応するシングルプレックスPCR | 34.00 | 20.46 | 25.19 |
| 103コピーのNFKBを用いるトリプレックスPCR | 35.17 | 19.94 | 28.38 |
| 対応するシングルプレックスPCR | 34.43 | 20.50 | 28.65 |
同じ反応中の参照遺伝子のコピー数が最大106倍多い場合でも、キットを用いれば、10コピー数程度の少数の標的遺伝子を検出できます(図「“ 過剰な参照遺伝子による10コピー標的遺伝子の検出」および対応する表「低存在量および高存在量の標的の定量的トリプレックスqPCRの成功」を参照)。
| CSBGの検出 | GAPDHの検出 | HSPの検出 | |
| テンプレートミックス1 | 1400コピー | 106コピー | 2 x 104コピー |
|---|---|---|---|
| トリプレックスPCRのCT値 | 27.73 | 18.69 | 23.59 |
| シングルプレックスPCRのCT値 | 27.08 | 18.89 | 23.52 |
| テンプレートミックス2 | 140 コピー | 106コピー | 2 x 103コピー |
| トリプレックスPCRのCT値 | 31.11 | 19.00 | 27.05 |
| シングルプレックスPCRのCT値 | 30.66 | 18.61 | 26.97 |
| テンプレートミックス3 | 14コピー | 106コピー | 2 x 102コピー |
| トリプレックスPCRのCT値 | 34.74 | 18.98 | 30.71 |
| シングルプレックスPCRのCT値 | 33.84 | 19.01 | 30.38 |
図参照
原理
QuantiTect SYBR Green PCR Kitsには、SYBR Green Iを使用してcDNA標的を高い特異性と感度でリアルタイム定量できるように最適化されたすぐに使用できるマスターミックスが含まれています(表“2x QuantiTect SYBR Green PCR Kitのコンポーネント”参照)。マスターミックスに含まれる蛍光色素SYBR Green Iは、数多くのさまざまな標的の分析を可能にし、標的特異的な標識プローブを合成する必要はありません。PCRバッファー中のK+イオンとNH4+イオンのバランスのとれた組み合わせは、特異的プライマーアニーリングを促進し、高いPCR特異性と感度を可能にします(図「特異的プライマーアニーリング」参照)。さらに、HotStarTaq DNA Polymeraseは厳密なホットスタートを提供し、非特異的産物の形成を防ぎます。
| コンポーネント | 特徴 | メリット |
| HotStarTaq DNA Polymerase | 95ºC 15分で活性化 | 室温でqPCR反応をセットアップ |
| QuantiTect SYBR Green PCR Buffer | NH4+ イオンと K+ イオンのバランスの取れた組み合わせ | 特異的なプライマーアニーリングにより信頼性の高いPCR結果を保証 |
| dNTPミックス | dTTPを部分的に置換し、オプションで反応のUNG処理を可能にするdUTPを含む | オプションのUNG処理により、PCR産物のキャリーオーバーによるコンタミネーションを排除 |
| SYBR Green I色素 | 二本鎖DNAと結合すると強い蛍光シグナルを発する | 高感度定量 |
| ROX色素 | Applied BiosystemsおよびAgilentの装置(オプション)での蛍光シグナルの正規化用 | ROX色素を必要とするサイクラーでの正確な定量。他のリアルタイムサイクラーの反応に干渉しない |
QuantiTect Probe PCR Kitsには、配列特異的プローブを使用してgDNAおよびcDNA標的を高い特異性と感度でリアルタイム定量できるように最適化されたすぐに使用できるマスターミックスが含まれています(表“2x QuantiTect Probe PCR Kitのコンポーネント”参照)。このキットは、加水分解プローブ検出(たとえば、TaqMan®やその他の二重標識プローブ)、FRETプローブ、Molecular Beaconを含むあらゆるタイプの配列特異的プローブに使用できるように設計されています。QuantiTect Probe PCR Kitsは、K+イオンとNH4+イオンのバランスの取れた組み合わせを含む独自のPCRバッファーを含んでおり、特異的プライマーアニーリングを促進し、高いPCR特異性と感度を可能にします(図「特異的プライマーアニーリング」参照)。さらに、HotStarTaq DNA Polymeraseは厳密なホットスタートを提供し、非特異的産物の形成を防ぎます。
| コンポーネント | 特徴 | メリット |
| HotStarTaq DNA Polymerase | 95ºC 15分で活性化 | 室温でqPCR反応をセットアップ |
| QuantiTect Probe PCR Buffer | NH4+ イオンと K+ イオンのバランスの取れた組み合わせ | 特異的なプライマーアニーリングにより信頼性の高いPCR結果を保証 |
| dNTPミックス | dTTPを部分的に置換し、オプションで反応のUNG処理を可能にするdUTPを含む | オプションのUNG処理により、PCR産物のキャリーオーバーによるコンタミネーションを排除 |
| ROX色素 | Applied BiosystemsおよびAgilentの装置(オプション)での蛍光シグナルの正規化用 | ROX色素を必要とするサイクラーでの正確な定量。他のリアルタイムサイクラーの反応に干渉しない |
QuantiTect Multiplex PCR Kitsは、マルチプレックスの2ステップRT-PCRを1回目の試行で成功させることができます(フローチャート「 QIAGENマルチプレックスキット」参照)。最適化されたマスターミックスは、マルチプレックス反応のPCR産物が、対応する単一増幅反応のPCR産物と同じ効率と感度で増幅されることを保証します。このキットを使用すると、わずか10コピーという少数の標的遺伝子を検出できます。
参照遺伝子とターゲット遺伝子を別々の反応ではなく、同じ反応で増幅すると、取り扱いエラーが最小限に抑えられ、遺伝子定量の信頼性が向上します。QuantiTect Multiplex PCR Master Mixは、K+イオンとNH4+イオンのバランスの取れた組み合わせと独自の合成 Factor MPを含み、これらはともにプライマーとプローブの核酸テンプレートへの安定した効率的なアニーリングを促進し、高いPCR効率を可能にします(表「2x QuantiTect Multiplex PCR Kitのコンポーネント」参照)。さらに、HotStarTaq DNA Polymeraseは厳密なホットスタートを提供し、非特異的産物の形成を防ぎます。
QuantiTect PCR KitsのマスターミックスにはdUTPも含まれているので、PCR開始前にウラシル-N-グリコシラーゼ(UNG)で前処理することができるため、確実にPCR産物のコンタミネーションがその後のPCR反応に影響を与えないようにすることができます。
| コンポーネント | 特徴 | メリット |
| HotStarTaq DNA Polymerase | 95ºC 15分で活性化 | 室温でqPCR反応をセットアップ |
| QuantiTect Multiplex PCR Buffer | NH4+ イオンと K+ イオンのバランスの取れた組み合わせ | 特異的なプライマーアニーリングにより信頼性の高いPCR結果を保証 |
| 合成Factor MP | 同じチューブで最大4つの遺伝子を高い信頼性でマルチプレックス分析 | |
| dNTPミックス | dTTPを部分的に置換し、オプションで反応のUNG処理を可能にするdUTPを含む | オプションのUNG処理により、PCR産物のキャリーオーバーによるコンタミネーションを排除 |
| ROX色素* | Applied BiosystemsおよびAgilentの装置(オプション)で蛍光シグナルを正規化 | ROX色素を必要とするサイクラーでの正確な定量。他のリアルタイムサイクラーの反応に干渉しない |
図参照
操作手順
必要に応じて、反応をウラシル-N-グリコシラーゼ(UNG)で前処理し、以前の反応からのPCR産物のキャリーオーバーを除去できます。リアルタイム2ステップRT-PCRで最適な結果を得るには、QuantiTect Reverse Transcription Kitを使用してcDNAを合成することをお勧めします。このキットは、ゲノムDNAのコンタミネーションを総合的に除去しながら、わずか20分でcDNAを高速合成します。
QuantiTect SYBR Green and Probe PCR Kitsは、面倒で時間がかかる可能性がある反応条件の最適化を必要としません。すぐに使えるPCRマスターミックスにプライマーとDNAテンプレートを加え、反応を開始させるだけです。ハンドブックのプロトコールに従って、あらゆるリアルタイムサイクラーで迅速に信頼性の高い結果が得られます。
QuantiTect SYBR Green PCR KitsをQuantiTect Primer Assaysと組み合わせて使用すると、遺伝子発現分析で非常に特異性の高い結果が保証されます。これらは、ヒト、マウス、ラット、その他数多くの種の転写産物を検出するための、ゲノムワイドでバイオインフォマティクス的に検証されたプライマーセットです。QuantiTect Primer Assaysは、GeneGlobeで簡単にオンライン注文できます。
QuantiTect Multiplex PCR Kitsには、反応条件やサイクリング条件を最適化する必要のないすぐに使えるマスターミックスが含まれています。ハンドブックには、利用可能なすべてのリアルタイムサイクラーで使用できる単一プロトコールが含まれており、推奨色素もリストされています。
キットはマスターミックス中にROXパッシブレファレンス色素を含むものと含まないものとがあり、実質的にどのリアルタイムサイクラーでも使用できます(表“適切なQuantiTect Multiplex PCR Kitの選択”参照)。ROX濃度が最適化されているため、低いコピー数であっても自動データ分析により検出が可能です。
| ROX色素 | キット | 互換性のあるサイクラー |
| マスターミックスに含まれる | QuantiTect Multiplex PCR Kit | Applied Biosystemsのサイクラー |
| マスターミックスに含まれない | QuantiTect Multiplex PCR NoROX Kit | Rotor-Geneサイクラー、およびBio-Rad、Cepheid、Eppendorf、Roche、Agilent、その他のサプライヤーのサイクラー |
アプリケーション
QuantiTect PCR Kitsは、あらゆるリアルタイムサイクラーでcDNAの遺伝子発現分析やgDNAの定量に使用できます。これにはApplied Biosystems、Bio-Rad、Cepheid、Eppendorf、Roche、Agilentの装置が含まれます。Rotor-Gene Qやその他のRotor-Geneサイクラーには、これら装置での高速サイクリング用に特別に開発されたRotor-Gene SYBR Green PCR Kit、Rotor-Gene Probe PCR Kit、またはRotor-Gene Multiplex PCR Kitの使用をお勧めします。
| 特徴 | QuantiTect SYBR Green PCR Kits | QuantiTect Probe PCR Kits | QuantiTect Multiplex PCR Kits |
| アプリケーション | ゲノムDNAまたはcDNA標的のリアルタイム定量 | ゲノムDNAまたはcDNA標的のリアルタイム定量 | ゲノムDNAまたはcDNA標的のリアルタイム定量 |
| 反応タイプ | PCRと2ステップRT-PCR | PCRと2ステップRT-PCR | マルチプレックスPCRとマルチプレックス2ステップRT-PCR |
| リアルタイムまたはエンドポイント | リアルタイム | リアルタイム | リアルタイム |
| サンプル/ターゲットタイプ | DNA、cDNA | DNA、cDNA | DNA、cDNA |
| シングルプレックスまたはマルチプレックス | シングル | シングル | マルチプレックス |
| SYBR Green Iまたは配列特異的プローブ | SYBR Green I | 配列特異的プローブ | 配列特異的プローブ |
| サーマルサイクラー | すべてのリアルタイムサイクラー(LightCycler, Rotor-Gene, ABIなど) | ほとんどのリアルタイムサイクラー(LightCycler、Rotor-Gene、ABIなど) | ほとんどのリアルタイムサイクラー(LightCycler、Rotor-Gene、ABIなど) |
| ROXあり/なし | ROXあり | ROXあり | ROXあり/なし |
裏付けデータと数値
広い直線範囲での特異的定量。