QIAquick Gel Extraction Kit
ゲル抽出あるいは酵素反応液からの最高10 µg までのDNA(70 bp~10 kb)クリーンアップ用
ゲル抽出あるいは酵素反応液からの最高10 µg までのDNA(70 bp~10 kb)クリーンアップ用
✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
Cat. No. / ID: 28704
✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
QIAquick Gel Extraction Kitは、スピンカラム、バッファー、コレクションチューブにより構成され、シリカメンブレンによりDNAフラグメントを400 mgまでのゲル切片あるいは酵素反応液から精製します。簡便で迅速な結合·洗浄·溶出ステップにより、70 bp~10 kbのDNAを30~50 µlの溶出液を用いて精製できます。pH指示薬入りバッファーにより、DNAがスピンカラムに結合する最適なpHを簡単にチェックできます。本キットはQIAcubeで完全自動化が可能です。
QIAquick Kitでは、高塩濃度バッファーによりDNAを結合し、低塩濃度バッファーあるいは水によりDNAを溶出するシリカゲルメンブレンを利用しています。この精製法でDNAサンプルからプライマー、ヌクレオチド、酵素、ミネラルオイル、塩、アガロース、臭化エチジウム、およびその他の夾雑物が除去できます(図 “ ゲルからの高回収率”)。シリカメンブレンテクノロジーにより樹脂漏れ、および懸濁液関連の問題や不便さは解消されます。それぞれの用途に応じて至適化された結合バッファーにより、種々の大きさのDNAフラグメントを選択的に吸着します。
GelPilot Loading Dye は3 種類のマーカー色素(xylene cyanol、bromophenol blueおよびorange G)が入っており、短いDNAフラグメントのゲルからの流出を回避でき、アガロースゲルの泳動時間の至適化が簡単に行なえます(図 " GelPilot Loading Dye")。
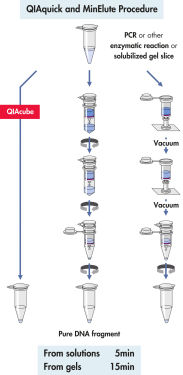
QIAquick Spin Column は2つの簡便な操作法オプションのためにデザインされました 。スピンカラムは簡便な卓上型マイクロ遠心機あるいはQIAvac 24 Plus のようなルアーコネクター付き吸引装置で使用できます。QIAquick Gel Extraction Kitは他のQIAGENスピンカラムを利用したキット同様にQIAcubeでの完全自動化が可能で、 標準化された再現性の高い結果が得られます(図 "Spin Column操作オプション A、 B、 C、 D、および E")。
QIAquickシステムで精製したDNAフラグメントは、シークエンシング、ライゲーション、トランスフォーメーション、制限酵素解析、標識反応、マイクロインジェクション、PCR、in vitro転写反応などのアプリケーションに即使用可能です。
| Features | Specifications |
|---|---|
| Binding capacity | 10 µg |
| Format | チューブ |
| Fragment size | 70 bp ~ 10 kb |
| Recovery: oligonucleotides dsDNA | 回収:dsDNAフラグメント |
| Processing | 手動 |
| Removal <10mers 17–40mers dye terminator proteins | 除去<10mers |
| Elution volume | 30 ~ 50 µl |
| Technology | シリカテクノロジー |
| Sample type: applications | DNA:PCR反応 |