✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
Cat. No. / ID: 28604
✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
特徴
- 非常に少量の溶出液量
- 迅速かつ簡単な操作
- 再現性と高い回収率
- サンプル操作を便利にするローディングダイ
製品詳細
MinElute Gel Extraction Kitには、最大400 mgのゲルから70 bp – 4 kb DNAフラグメント精製にシリカメンブレンスピンカラム、バッファーおよびコレクションチューブが含まれます。スピンカラムは、非常に少量(10 µL)の溶出で濃縮DNAを高収率で得られるよう設計されています。pH指示薬により、スピンカラムへのDNA結合の至適pHを容易に確認できます。MinEluteで精製されたDNAフラグメントは、シークエンシング、マイクロアレイ解析、ライゲーションと形質転換、制限酵素消化、標識、マイクロインジェクション、PCR、in vitro転写など様々なアプリケーションに直接使用できます。MinElute Gel Extraction Kitは、QIAcube Connectで自動化できます。
最適な結果を得るには、本製品をQIAvac 24 Plusと併用することをお勧めします。
パフォーマンス
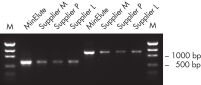
MinElute Gel Extraction Kitは、DNAサンプルからヌクレオチド、酵素、塩、アガロース、臭化エチジウムなどの不純物を除去し、ダウンストリームのさまざまなアプリケーションへの高濃縮DNAが得られます(「 高濃縮DNA」の図を参照)。
MinElute Gel Extraction Kitは、ゲル抽出用のスピンカラムキットです。小型遠心機または真空マニホールドを使用して、70 bp – 4 kbの高濃度の DNA を迅速に精製します。(4 kb – 10 kbのDNAフラグメントはQIAquick Gel Extraction Kitで、また70 bpより小さいまたは10 kbより大きいDNAフラグメントはQIAEX II Gel Extraction Systemで精製してください。)
図参照
原理
MinElute Kitsは、高塩濃度バッファーでDNAを結合し、低塩濃度バッファーまたは水で溶出するためのシリカメンブレンキットです。シリカメンブレン技術は、緩い樹脂やスラリーによる問題や不都合がありません。特殊な結合バッファーは、特定のアプリケーションに最適化され、特定のサイズ内のDNA分子の選択的な吸着を促進します。
Gel loading dye
ローディングダイは、迅速で便利なサンプル処理と分析を可能にします。GelPilot Loading Dyeは、3種類のトラッキングダイ(キシレンシアノール、ブロモフェノールブルー、およびオレンジG)を含み、アガロールゲルのランタイムの最適化を容易にし、より小さなDNAフラグメントの過度な移動を防ぎます(「 GelPilot Loading Dye」の図を参照)。
図参照
操作手順
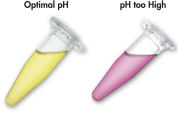
MinEluteシステムは、シンプルな結合-洗浄-溶出の操作です(「 MinEluteの操作手順」のフローチャートを参照)。ゲルスライスは、pH指示薬を含むバッファーに溶解するので、DNA結合の至適pHを決定でき(「 pH Indicator Dye」の図を参照)、その混合液をMinEluteスピンカラムにアプライします。核酸は、バッファーによる高塩濃度の条件下でシリカゲルメンブレンに吸着します。不純物は洗い流され、純粋なDNAは、少量の低塩濃度バッファーまたは水と共に溶出し、後のアプリケーションに使用できます。
取り扱い
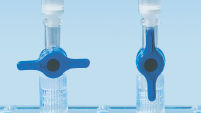
MinEluteスピンカラムは、2通りの操作方法が設定されてます。スピンカラムは、小型遠心機または QIAvac Luer Adapters付きのQIAvac 24 Plusなどのルアーコネクター付きの真空マニホールド上にフィットします。MinElute Gel Extraction Kitは、その他のQIAGENスピンカラムキットに加えて、QIAcube Connectで 完全自動化して、生産性の向上、結果の標準化することができます (「スピンカラムの取り扱いオプション A、 B、 C、 D および QIAcube Connect」の図を参照)。
図参照
アプリケーション
MinEluteまたはQIAquick Systemで精製したDNAフラグメントは、以下を含むすべてのアプリケーションで使用できます。
- 次世代シークエンシングを含むシークエンシング
- マイクロアレイ解析
- ライゲーションと形質転換
- 制限酵素処理
- Labeling
裏付けデータと数値
スピンカラム取り扱いオプション — C。

Specifications
| Features | Specifications |
|---|---|
| Binding capacity | 5 µg |
| Elution volume | 10 µl |
| Fragment size | 70 bp ~ 4 kb |
| Sample type: applications | DNA:PCR反応 |
| Technology | ゲル抽出 |
| Recovery: oligonucleotides dsDNA | 回収:dsDNAフラグメント |
| Format | チューブ |
| Processing | 手動 |