Products
特徴
- 分子バーコード(UMI)による正確な定量とシークエンス解析
- GeneGlobe によるクラウドベースの無償データ解析ツールを提供
- 精製用のQIAseq 磁気ビーズが同梱
- ヒトまたはマウスサンプルの自動化に適したプロトコール
- サンプルインデックスで最大384 サンプルまでマルチプレックス可能
製品詳細
QIAseq Immune Repertoire RNA Library Kitは、分子バーコード(UMI)および遺伝子特異的プライマーを使用したRNAターゲットシークエンシング用ライブラリー調製キットです。ヒトおよびマウスT細胞受容体パネルは、T細胞レセプター(TCR)レパトアのCDR1、CDR2およびCDR3領域を含む、4つの鎖(アルファ、ベータ、ガンマおよびデルタ)を全て解析可能です。
クラウド上のGeneGlobe Data Analysis Centerを介した無償データ解析ツールにより、最初のリードマッピング、分子バーコード(UMI)毎のデータ分配から、T細胞受容体の各アルファ、ベータ、ガンマおよびデルタ鎖毎のデータ、CDR3ペプチド配列や長さの分布、rarefraction解析(希釈法)による多様性解析およびV/D/J 領域を使用したヒートマップが出力されます。
パフォーマンス
Comprehensive view of the T-cell immune repertoire
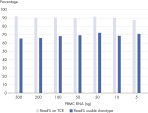
The heatmaps allow for easy identification of enriched clonotypes across the sample. This figure shows the major clonotype of the Jurkat cell, as well as the diversity of the PBMC background. The data analysis included with the purchase of the QIAseq Immune Repertoire T-cell receptor panels includes an online portal that seamlessly integrates with Illumina BaseSpace and provides primary read mapping, UMI demultiplexing and reports on sequencing performance, TCR chain usage, CDR3 peptide sequence and length distributions, together with rarefaction and V/D/J usage heat maps.Sensitive to at least 0.01%
RNA from Jurkat cells was spiked into RNA extracted from peripheral blood mononuclear cells (PBMCs; Precision Medicine) at 10%, 1%, 0.1% and 0.01% and used to make an RNA-seq library. Table 1 shows the number of raw reads and the demultiplexed unique captures (UMIs) per Jurkat TCR-alpha and TCR-beta clonotype. Even when present at only 0.01%, the Jurkat RNA is readily quantifiably identified. For data analysis, UMIs and Raw Reads are used to ensure high precision around each clonotype sequence identified.
| Chain | % Jurkat cells | Rank | Reads | UMIs |
|---|---|---|---|---|
| TCR-alpha | 10 | 1 | 751,749 | 107,150 |
| 1 | 1 | 146,959 | 20,692 | |
| 0.1 | 1 | 10,708 | 1,742 | |
| 0.01 | 10 | 1,306 | 217 | |
| TCR-beta | 10 | 1 | 383,594 | 40,943 |
| 1 | 1 | 5,920 | 7,541 | |
| 0.1 | 2 | 5,401 | 620 | |
| 0.01 | 61 | 457 | 60 |
図参照
原理
The QIAseq Immune Repertoire RNA Library Kit relies on a highly efficient, TCR-specific cDNA synthesis reaction, ligation of sample index adapters containing UMIs and TCR gene-specific primer enrichment for sensitive TCR clonotype and diversity assessment. Each kit contains species-specific TCR reverse transcriptase and enrichment panel primers, together with QIAseq reaction cleanup beads and library reagents. The QIAseq Immune Repertoire RNA Library Kit is designed to enrich TCR α, β, γ and σ subunits using 10–1000 ng RNA from human or mouse samples.
操作手順
The QIAseq Immune Repertoire RNA Library Kit relies on a highly efficient, TCR-specific cDNA synthesis, TCR gene-specific primer enrichment and molecular indexing for accurate and sensitive TCR clonotype and diversity assessment (see figure " QIAseq Immune Repertoire RNA Library workflow"). TCR reverse transcriptase and enrichment panel primers are provided, together with library reagents.
cDNA synthesis
RNA samples are first reverse transcribed into cDNA with TCR-specific RT primers. Subsequently, second-strand synthesis occurs, which generates double-stranded cDNA (ds-cDNA). This ds-cDNA is then end-repaired and A-tailed in a single-tube protocol.
UMI assignment
Prior to target enrichment and library amplification, each original cDNA molecule is assigned a UMI by ligating an adapter containing a 12-base fully random sequence (i.e., the UMI) to the ds-cDNA. Statistically, this process provides 4^12 possible indices per adapter, and each DNA molecule in the sample receives a unique UMI sequence. In addition, this ligated adapter also contains the first sample index.
Target enrichment and final library construction
Following UMI assignment, target enrichment is performed to ensure that TCR cDNA molecules are sufficiently enriched in the sequenced library. For enrichment, ligated cDNA molecules are subjected to targeted PCR using one TCR constant-region-specific primer and one universal primer complementary to the adapter. A universal PCR is ultimately carried out to amplify the library and introduce platform-specific adapter sequences, as well as additional sample indices.
図参照
アプリケーション
裏付けデータと数値
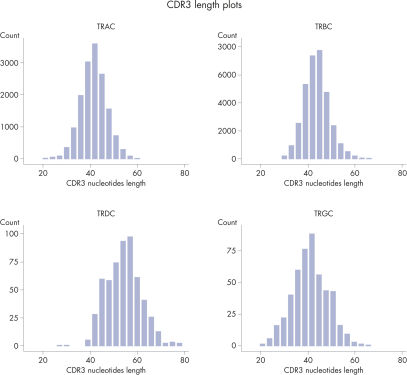
CDR3 Length Plots are Shown for Each Receptor in from a Single Sample.