Taq DNA Polymerase (250 U)
Cat. No. / ID: 201203
特徴
- PCR条件の至適化が最小限で済むQIAGEN PCR Buffer
- 直接ゲルにロード可能なPCR バッファーでより迅速な操作
- GC リッチなテンプレートの増幅を促進するQ-Solution
- 簡便で取り扱いやすいフォーマット
製品詳細
パフォーマンス
Taq DNA Polymeraseは、他メーカーの試験されたキットに比べて優れており、時間のかかる至適化を必要とせずに、様々なPCR条件で確固たるPCR性能を示します(図" Tm 値の異なるプライマーへの適応性" と" 長いPCR産物の特異的増幅")。TaqDNA Polymeraseは、全てのロットで、低コピー数のヒトゲノムDNAをターゲットとした増幅実験により、PCR特異性の厳密さや再現性をチェックするなど、広範囲な品質管理テストを受けています(図 " ロット間での再現性")。同様にキットに含まれているQIAGEN PCR BufferとCoralLoad PCR Bufferのユニークな組成によって、最小限の至適化に必要な事項と様々なPCR条件で、特異性の高いPCRが可能となります(図" 幅広い至適アニーリング温度"と " 異なったマグネシウム濃度への適応")。さらに、CoralLoad PCR Bufferは、PCR産物をアガロースゲルに即座にロードすることを可能にするため、比較的簡単な操作で比較的早く結果が得られます。また、キット付属のPCR 添加物、Q-Solutionを使用すれば、不適切なPCR条件を改善することができます(図 " 増幅困難なテンプレートの増幅")。
Taq DNA Polymeraseの詳細データ
濃縮:5 units/µl
組み換え酵素:はい
基質類似体: dNTP、ddNTP、dUTP、ビオチン-11-dUTP、DIG-11-dUTP、蛍光-dNTP/ddNTP
エクステンション率72℃ で2~4 kb/分
半減期:97℃で10分、94℃で60分
増幅効率:≥105 倍
5'->3' エキソヌクレアーゼ活性:あり
余分なA付加:あり
3'->5' エキソヌクレアーゼ活性:なし
混入しているヌクレアーゼ:なし
混入しているRNase:なし
混入しているプロテアーゼ:なし
自己プライミング活性:なし
図参照
原理
Taq DNA Polymerase は、スタンダードなPCRから特殊なPCRまで適応できる 高品質の組み換え酵素です(図 " Tm 値の異なるプライマーへの適応性" と" 長いPCR産物の特異的増幅")。
QIAGEN PCR Buffer
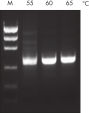
革新的なQIAGEN PCR Bufferは、PCR至適化の操作を軽減することにより、時間と労力を節約するために開発されました。QIAGEN PCR Bufferは、KCl も (NH4)2SO4 も含んでいます(図" プライマーのアニーリングにおける特異性が増加")。このユニークなバッファーによって、特異的PCRの産物を増幅しやすくなります。PCR サイクルごとのアニーリングの間、非特異的なプライマー結合に対する特異的な結合の比率が、バッファーによって高く保たれます。このPCRバッファーは、KClと (NH4)2SO4 のユニークな配合比により、従来のPCR バッファーに比べ幅広いアニーリング温度やMg2+濃度の範囲で厳密なプライマーアニーリング条件を実現します。異なるアニーリング温度あるいはMg2+ 濃度を用いて行なうPCRの至適化は、劇的に緩和され、多くの場合、不要となります(図 " 幅広い至適アニーリング温度"および" 異なったマグネシウム濃度への適応 ")。
CoralLoad PCR Buffer
CoralLoad PCR Bufferは、QIAGEN PCR Bufferの全ての特長を持っています。さらに、PCR反応液をアガロースゲルに直接ロードするために使用することができ、従って、ゲルローディングバッファーを別途加える必要はありません。CoralLoad PCR Bufferは従来のQIAGEN PCR Bufferと同程度の高いPCR特異性を持ち、反応の至適化は最小限に抑えられます。さらに、これに入っている2種類のマーカー色素(オレンジ色の色素と赤色の色素)により、DNAの移動距離の予測とアガロースゲルの泳動時間の至適化を容易に行なえます(図" CoralLoad PCR Buffer")。本バッファーはピペッティングの目視化を改良し、PCR産物をゲルに直接ロードすることが可能なので、簡便性が増加します。
Q-Solution
Q-Solution は、DNAの溶解挙動を変えることで、GCリッチなテンプレート、または二次構造が複雑なテンプレートを増幅しやすくします。この試薬の添加により、しばしば最適条件ではないPCRが改善されたり、今まで得られなかったPCR産物の増幅が可能になったりします (図 " 増幅困難なテンプレートをQ-Solutionにより増幅")。DMSO や他のPCR添加物と異なり、本溶液はどのようなプライマー/テンプレートシステムでも一定の濃度で作用し、毒性もありません。
図参照
 Specific amplification of long PCR products.
Specific amplification of long PCR products.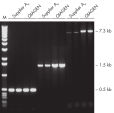 NH4+ and K+ cations in QIAGEN PCR buffers increase specific primer annealing.
NH4+ and K+ cations in QIAGEN PCR buffers increase specific primer annealing.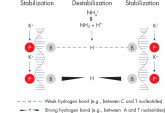 A. Wide annealing temperature window. B. Tolerance of variable magnesium concentration.
A. Wide annealing temperature window. B. Tolerance of variable magnesium concentration.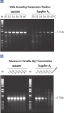 CoralLoad PCR Buffer.
CoralLoad PCR Buffer.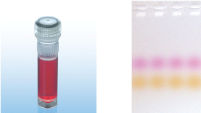 Amplification of difficult templates with Q-Solution.
Amplification of difficult templates with Q-Solution.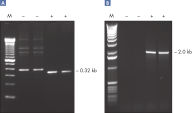
操作手順
アプリケーション
TaqDNA Polymeraseは以下のようなスタンダードな用途にも特殊な用途にも使用できます。
- スタンダードな PCR
- RT-PCR
- スクリーニング
- PCRによるDNAフィンガープリンティング(VNTR、STR、RAPD)
裏付けデータと数値
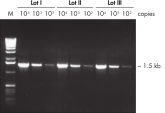
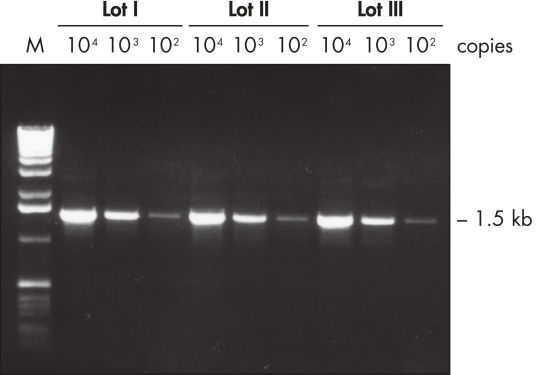
Lot-to-lot reproducibility.
A fragment of the single-copy gene for cystic fibrosis was amplified from 30 ng, 3 ng, and 300 pg human genomic DNA corresponding to 104, 103, and 102 copies of target template, respectively. Three different lots of QIAGEN Taq DNA Polymerase were used and equal volumes of the PCR product were analyzed on a 1% agarose gel. M: markers.
Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, RT-PCR, DNA fingerprinting |
| dNTP's included | No |
| Real-time or endpoint | Endpoint |
| Reaction type | PCR amplification |
| Single or multiplex | Single |
| With/without hotstart | Without hotstart |
| Enzyme activity | 5' -> 3' exonuclease activity |
| Mastermix | No |
| Sample/target type | Genomic DNA and cDNA |