QIAseq Targeted Methyl Panel (8)
Cat. No. / ID: 335501
特徴
- わずか1日でDNAのバイサルファイト変換からシークエンスライブラリー構築を実現するワークフロー
- 分子バーコード(UMI)による正確なメチル化定量解析
- Single-Primer Extension(SPE)技術により困難な配列へのプライマーデザインも可能
- GeneGlobe によるクラウドベースのフリーのデータ解析ツール、もしくはCLC Genomics Workbenchソフトウェアによりデータ解析までサポート
製品詳細
QIAseq Targeted Methyl Panelsは、わずか一日のワークフローで、目的領域のメチル化解析を実現します。Single-Primer Extension (SPE) 技術に基づくプライマーデザインにより、これまで困難であったCpGメチル化ターゲットへのカバレッジを最大化し、分子バーコード(UMI)による高精度なメチル化定量解析を可能にします。
本製品のカスタムデザインをご希望の場合は、QIAseq Targeted Methyl Custom Panels をご覧ください。
パフォーマンス
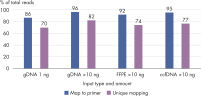
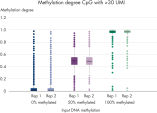
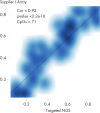
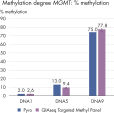
QIAseq Targeted Methyl Panels deliver high mapping efficiency (figure QIAseq Targeted Methyl Panel: mapping efficiency), high reproducibility (figure QIAseq Targeted Methyl Panel: methylation degree reproducibility) and high correlation with established methods (figures QIAseq Targeted Methyl Panel – high correlation with established methods and QIAseq Targeted Methyl Panel: methylation status of FFPE DNA: methylation degree).
図参照
原理
Epigenetics describes the study of heritable changes in gene function that occur without a change in the nuclear DNA sequence. In addition to RNA-associated silencing and histone modification, a major epigenetic mechanism in higher-order eukaryotes is DNA methylation. Epigenetic changes play a crucial role in the regulation of important cellular processes, such as gene expression and cellular differentiation, and were also identified as key factors in various diseases.
DNA methylation occurs on cytosine residues, especially in CpG islands, which are GC-rich regions. They are usually clustered around the regulatory region of genes and can affect their transcriptional regulation. Methylation of CpG islands is known to inactivate gene expression and plays an important role in normal and disease development. Cytosine methylation may also occur in non-CpG content, as described for embryonic stem cells.
QIAseq Targeted Methyl Panels enable Sample to Insight, targeted next-generation sequencing (NGS) to interrogate DNA methylation degree. This highly optimized solution facilitates sensitive DNA methylation detection using integrated Unique Molecular Indices (UMIs) from cells, tissues and biofluids. The required amount of template for a single QIAseq Targeted Methyl sequencing reaction ranges from 1–100 ng for fresh gDNA, 10–200 ng for FFPE DNA or 10–100 ng for ccfDNA.
操作手順
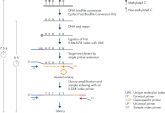
QIAseq Targeted Methyl Panels employ a streamlined workflow (figure QIAseq Targeted Methyl Panel sequencing workflow) for generating focused libraries from multiple input types, including difficult samples such as FFPE and liquid biopsy (ccfDNA). First, bisulfite conversion of isolated DNA is performed (recommended product: EpiTect Fast Bisulfite Kit), followed by DNA end repair. Afterwards, during target enrichment, a Unique Molecular Index (UMI) is added to the DNA library. UMIs are unique barcode sequences that enable bioinformatics analysis to exclude duplicates and error-correct the sample’s methylation status. QIAseq Targeted Methyl Panels utilize the QIAseq enrichment technology, a PCR method that uses a single target-specific primer, with a universal primer. This approach increases the efficiency of design – making it more sensitive for challenging samples, while maximizing the coverage of the regions of interest.
図参照
アプリケーション
- Detection of methylation status from FFPE samples
- Detection of methylation status from liquid biopsy
- Detection of cell- and tissue-specific methylation markers
裏付けデータと数値
QIAseq Targeted Methyl Panel – high correlation with established methods