RNeasy FFPE Kit (50)
Cat. No. / ID: 73504
特徴
- ホルマリンによるクロスリンクを切断する新しい手法
- RNA分解がなく効率的にRNAを遊離
- わずか85 分でRNA が得られる能率化されたプロトコール
製品詳細
RNeasy FFPE Kitはホルマリン固定、パラフィン包埋組織切片からのトータルRNA精製用に特別にデザインされています。特別な溶解およびインキュベーション条件によりホルムアルデヒドによるクロスリンクを元に戻します。さらに溶解バッファーは、組織切片からRNAを効率的に遊離し、操作におけるRNA分解もありません。また本キットでは、混入したゲノムDNAコンタミを最適に除去するためにDNaseとDNase Booster Bufferを使用しています。また、わずか14 μl の溶出容量でのトータルRNA 精製が可能なRNeasy MinElute スピンカラムを使用しています。精製はQIAcube上で全自動化できます。
パフォーマンス
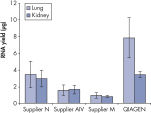
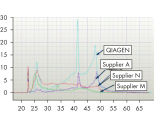
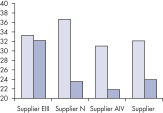
図参照
原理
ホルマリン固定された組織はRNA–RNAおよびRNA–タンパク質間のクロスリンクが生じているため、酵素によるアッセイ系でのRNAの使用において良好な結果が得られません。FFPEサンプルでは、固定および包埋条件も核酸の過度なフラグメント化をもたらします。RNeasy FFPE Kitは、特別な溶解およびインキュベーション条件によりホルムアルデヒドによるクロスリンクを元に戻します。溶解バッファーはFFPE組織サンプルからRNAを効率的に遊離させ、RNA分解を回避します。これらの至適条件により、全ての使用可能なRNAの精製が可能になるため、RNeasy FFPE KitでFFPEサンプルから精製されるRNA収量は、ほかの精製法を用いたものよりも収量よりも大きくなります。
操作手順
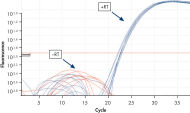
図参照
アプリケーション
RNeasy FFPE Kitは、70 ヌクレオチドより長い全てのRNAを分離するので、RT-PCRなどの多数のアプリケーションに有用なRNAフラグメントを回収できます。しかし、FFPEのサンプルから精製されたRNAはフラグメント化が激しく、完全長RNAを必要とするダウンストリームアプリケーションには使用するべきではありません。いくつかのアプリケーションではフラグメント化したRNAを使用できるように変更が必要な場合があります(例:RT-PCRで短い増幅産物にするようにプライマーをデザインする)。cDNA合成にはオリゴ-dTプライマーの代わりにランダムプライマーか遺伝子特異的なプライマーを使用するべきです。
裏付けデータと数値
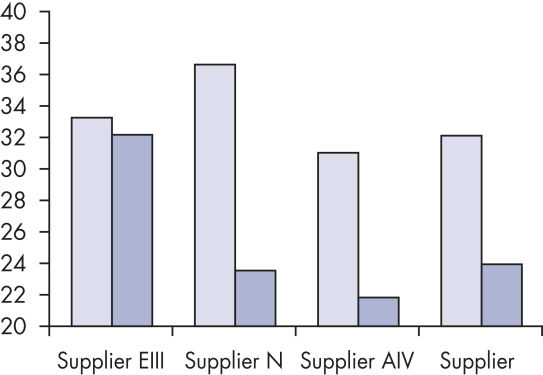
Reliable results in real-time RT-PCR analysis.
Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, qPCR, real-time RT-PCR, microarray |
| Elution volume | 14–30 µl |
| Main sample type | FFPE tissue samples |
| Processing | Manual (centrifugation), automated (QIAcube) |
| Format | Spin column |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein | RNA |
| Time per run or per prep | 70 minutes |
| Sample amount | 1*5 µm to 4*10µm sections |
| Technology | Silica technology |
| Yield | Varies |