✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
Rotor-Gene Q 2plex Priority Package Plus
Cat. No. / ID: 9001862
✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
特徴
- 遠心エアコントロール方式でサンプル間の高い温度均一性を実現(±0.02℃)
- UVから赤外線領域までをカバーする広範囲にわたる光学システムに対応
- 使いやすいソフトウェアで最新の解析をサポート
- 適応性の高いデザインにより最高の簡便性と最低限のメンテナンス
- QIAGENアッセイと併用し各種アプリケーションで良好な結果を実現
製品詳細
QIAGEN のリアルタイムPCR 装置Rotor-Gene Qは、最高のパフォーマンスを実現するために数多くの工夫がなされており、研究ニーズにマッチした最適な結果を実現します。また、リアルタイムPCR用に至適化済みのQIAGENキットと組み合わせることにより、Rotor-Gene Q は様々なアプリケーションに効率よく対応することができます。
パフォーマンス
図参照
原理
遠心エアコントロール方式による卓越した性能
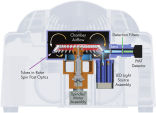
遠心エアコントロール方式を採用した Rotor-Gene Q は、市販されているこれまでのリアルタイムPCR装置の中で、最も精密性と機能性に富んでいます(図 " Rotor-Gene Q の断面図")。遠心エアコントロール方式では、サンプルローターが常に遠心されていることにより、反応サンプル間の温度均一性を高く保持します(±0.02 ℃)。どのサンプルも同様に均一に検出全てのサンプルは回転しながら光源/検出器間の光路上を通過し、光源/サンプル/検出器の一定した短い光学的距離でその蛍光シグナルを迅速に得ることができます。このように温度および光学的に均一であるため、感度の高い正確かつ迅速なリアルタイムPCR解析結果が得られ(図 " 正確なリアルタイムPCR 解析")、またサンプル間のばらつきやエッジ効果をなくします。これらは、従来のブロックベースの機器では、ブロック全体の温度勾配や複数の複雑な光経路のため避けられない問題でした。
遠心エアコントロール方式の特長:
チューブ間のばらつきは ±0.02℃
均一な検出により、ROX のようなリファレンス色素が不要
高速な加熱・冷却機構によるランニング時間の短縮
高い感度と再現性
広い光源領域で様々なアプリケーションを実現
Rotor-Gene Q は、SYBR® Green のようなインターカレーター色素や、加水分解プローブ(TaqMan プローブ)、ハイブリダイゼーションプローブ(FRET プローブ)、Scorpion プローブ、その他幅広い蛍光波長の検出に最適です。搭載波長は、UVから赤外波長まで、最高6 チャンネルを搭載しています。さらに、ソフトウェア上での簡単な設定により、それぞれの励起光源/検出フィルターの組み合わせをカスタムで作製できるので、特殊な波長の色素にもRotor-Gene Q は対応可能です。
| 蛍光検出用チャンネルチャンネル励起波長 (nm) | 検出波長 (nm) | 検出可能な蛍光色素の例 | ||
| Blue | 365 ± 20 | 460 ± 20 | Marina Blue、Edans、Bothell Blue、 | Alexa Fluor 350、AMCA-X |
| Green | 470 ± 10 | 510 ± 5 | FAM、SYBR® Green I、Fluorescein、 | EvaGreen、Alexa Fluor 488 |
| Yellow | 530 ± 5 | 557 ± 5 | JOE、VIC、HEX、TET、MAX、CAL Fluor Gold 540、Yakima Yellow | |
| Orange | 585 ± 5 | 610 ± 5 | ROX、CAL Fluor Red 610、Cy 3.5、 | Texas Red、Alexa Fluor 568 |
| Red | 625 ± 5 | 660 ± 10 | Cyanine 670、Quasar 670、LightCycler | Red640、Alexa Fluor 633 |
| Crimson | 680 ± 5 | 712 高域 | Quasar 705、LightCycler Red705、 | Alexa Fluor 680 |
| HRM | 460 ± 20 | 510 ± 5 | SYBR® Green I、SYTO9、LC Green、 | LC Green Plus+、EvaGreen |
HRM 解析によるPCR 産物の識別
High Resolution Melt(HRM)解析とは、融解曲線解析を極めて精度の高い温度分解能を利用して、わずかなTm 値の差を検出することができる技術です。HRM法は、二本鎖PCR産物を解離(融解)性に基づいて解析します。古典的な融解曲線解析に類似していますが、幅広いアプリケーション用にさらに多くの情報が得られます。PCR産物は、その塩基組成(GC 含量)、配列、長さなどに依存した産物特有の解離温度(Tm 値)を有します。Rotor-Gene Q の温度分解能は±0.02 ℃であるため、PCR 産物中に1塩基の違いがあっても、高感度に検出することができます。Rotor-Gene Q の遠心エアコントロール方式による卓越した温度均一性と光学的均一性はHRM に最適です。
Rotor-Gene Q のHRM 機能の特長:
専用のHRM 励起チャンネルを搭載
±0.02 ℃の超高温度分解能による厳密な識別
高速データ収集
自動ジェノタイピング機能を搭載したソフトウェア
最小限のメンテナンスで最大の簡便さ
Rotor-Gene Q は、日常のメンテナンスが必要なく、使いやすい設計になっています。そのため、装置本体のメンテナンスにかける時間を低減し、研究に集中することが可能です。
Rotor-Gene Q の特長:
非常に安定したLED使用のため、高価なランプやレーザーの交換が不要で、光源性能が徐々に低下することが少ない
設置時や機器移動時に必要とされる光学的補正は不要
ブロック型のような清掃が不要
回転により反応中の結露や気泡が発生しない
小さくて軽い装置なので使いやすく、ラボのどこにでも設置可能
図参照
操作手順
スループットに応じて変更できる多様なフォーマット
Rotor-Gene Q は多様なニーズに対応するため、様々なPCR チューブフォーマットを使用できます(図 " 多様なフォーマット")。チューブを搭載するローターを取り替えるだけでフォーマットをわずか数秒で変更できます。よりスループットの高い実験系には、Rotor-Disc が使用できます。Rotor-Disc は、円周上にサンプルウェルが配置された円形プレートです。Rotor-Disc 100を用いることで、96サンプルプレート処理と同等のスループットを実現することができ、残りの4ウェルはコントロール反応などに使用することができます。余ったウェルは、追加の反応や追加コントロールに便利に使用することができます。72ウェルのRotor-Disc 72 もあります。Rotor-Disc は、Rotor-Disc Heat Sealer を用いてプラスチックフィルムで簡単かつ迅速にシールできます。Rotor-Disc を使用する反応で必要なものは全て Rotor-Disc 100 Starter Kit あるいはRotor-Disc 72 Starter Kit に入っています。
マニュアルで反応セットアップを行なうことも、QIAGENの自動化ソリューションであるQIAgiliyを利用して迅速かつ高精度のPCRセットアップを行なうこともできます。また、QIAsymphony ASは、日常的にルーチンのPCRテストを行なうラボに最適です。どちらの装置もRotor-Gene フォーマットの自動反応セットアップを行なうことができ、サンプルリストの直接転送が可能で、リアルタイムPCRマスターミックス用の検証済みプロトコールが付属しています。
装置の定期的なバリデーションも実施可能
検査機関や診断を目的とした研究施設では、使用しているサーマルサイクラーの温度精度のバリデーションを定期的に実施することを求められる場合があります。Rotor-Gene Q では、温度精度の自動キャリブレーションを行なう専用ローター"Rotor-Disc OTV(Optical Temperature Verification)Kit"をセットしてランニングするだけで、自動的に装置の温度キャリブレーションを実施することができます。全操作は、わずか数分なので、ほかの装置のようなサービスエンジニアの出向も要請する必要もなく、費用対効果も高く効率的です。
図参照
アプリケーション
数々のRotor-Gene Q用QIAGEN キットは、反応条件やサイクル条件の至適化の必要がなく、リアルタイムPCR アプリケーションにおいて信頼できる定量を実現します。以下の分野におけるリアルタイムPCR およびHRMに特化したキットを販売しています。
- 遺伝子発現解析
- 病原体検出
- DNA メチル化解析
- ジェノタイピングおよび遺伝子スキャニング
- miRNA 研究
ソフトウェア
定量化を可能にし、データの信頼性を向上するソフトウェア
この包括的なRotor-Gene Q software packageは、基本的から高度なアルゴリズムを搭載しており現在最高水準のリアルタイムPCR解析手順をサポートします。これにより、お客様の価値ある実験データ解析の自由度と結果に対する信頼性が増します。データはランを開始してから結果を出力するまでの全てのプロセスのステップをトラッキングできることで確実になります。
HRM解析を用いたジェノタイピングおよび変異検出用の優れたソフトウェア
Rotor-Gene ScreenClust HRM SoftwareはRotor-Gene operating softwareの拡張版です。このソフトウェアはRotor-Gene QまたはRotor-Gene 6000サイクラーから得られたHRMデータの解析のために現在使用されている最も強力なツールです。サンプルをクラスタごとに分類することで、Rotor-Gene ScreenClust HRM Softwareはジェノタイピングおよび変異スクリーニングのようなアプリケーション用のHRM解析の新しい局面を展開します。
サービス
Have your throughput demands increased? Do you want to expand your application range or further streamline workflows? Do you need to manage more complex samples? Do you need improved connectivity?
If the answer to any of these questions is yes, then QIAGEN’s trade-in/trade-up program is just right for you!
QIAGEN makes it easy to keep up to speed with the latest automation and sample processing technologies. Simply exchange an old instrument for a new one. Improve your efficiency by trading in a competitor instrument or trading up with an older QIAGEN instrument.
裏付けデータと数値
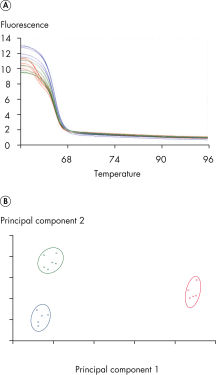
Identification of a class IV SNP.
Specifications
| Features | Specifications |
|---|---|
| 寸法 | Width, 37 cm (14.6 in.); Height, 28.6 cm (11.3 in.); Depth (without cables), 42 cm (16.5 in.); Depth (door open), 53.8 cm (21.2 in.) |
| Operating temperature | 18-30ºC (64-86ºF) |
| Pollution level | 2; Environmental class 3K2 (IEC 60721-3-3) |
| Humidity | 10–75% (noncondensing) |
| Features | Dynamic range, 10 orders of magnitude |
| Protocols/main application on this instrument | Gene expression analysis, microRNA detection, Virus detection, SNP genotyping, SNP genotyping, High Resolution Melt analysis (HRM) |
| ソフトウェア | Q-Rex Software, supplied on the installation CD provided |
| Overvoltage category | II |
| Heat dissipation/thermal load | Average, 0.183 kW (632 BTU/hour); Peak, 0.458 kW (1578 BTU/hour) |
| Optical System | Up to 6 channels spanning UV to infra-red wavelengths; Excitation sources: High energy light-emitting diodes; Detector: Photomultiplier; Acquisition time: 4 s. The software allows to create new excitation/detection wavelength combinations |
| Power | 100–240 V AC, 50–60 Hz, <520 VA (peak); Power consumption <60 VA (standby); Mains supply voltage fluctuations are not to exceed 10% of the nominal supply voltages; F5a 250 V fuse |
| Kits designed for this instrument | artus QS-RGQ Kits (not available in all countries), RG SYBR Green PCR Kits; RG SYBR Green RT-PCR Kit; RG Probe PCR Kits; RG Probe RT-PCR Kit; RG Multiplex PCR Kit |
| Place of operation | For indoor use only |
| Samples per run (throughput) | Tubes 0.2 ml; Strip Tubes 0.1 ml (4 tubes); Rotor-Disc 72; Rotor-Disc 100; Up to 100 samples per run using a Rotor-Disc 100 |
| Storage conditions | 15ºC to 30ºC (59ºF to 86ºF) in manufacturer’s package; Max. 75% relative humidity (noncondensing); Environmental class 1K2 (IEC 60721-3-1) |
| Technology | Real-time PCR cycler |
| Transportation conditions | –25ºC to 60ºC (–13ºF to 140ºF) in manufacturer’s package; Max. 75% relative humidity (noncondensing); Environmental class 2K2 (IEC 60721-3-2) |
| Warranty | 1 year on instrument; lifetime guarantee on excitation LEDs |
| Thermal performance | Temperature range, 35 to 99ºC; Temperature accuracy, ±0.5ºC (type, measured 30 seconds after clock start); Temperature resolution, ±0.02ºC (smallest programmable increment); Temperature uniformity, ±0.02ºC |
| Typical run time | 40 cycles in 45 min (assay dependent) |
| 重量 | 12.5 kg (27.6 lb.), standard configuration |
| Altitude | Up to 2000 m (6500 ft) |
サービスプラン
RGQ, Full Agreement
Cat. No. / ID: 9242698
RGQ, Full Agreement, no PM
Cat. No. / ID: 9241781
RGQ, Core Agreement
Cat. No. / ID: 9245253