GeneGlobeで探索・設定する
適切なターゲット特異的アッセイおよびパネルを探すか、またはターゲットをカスタムデザインし、お客様のご興味のある生物学的ターゲット評価にお使いください。
qBiomarker Copy Number PCR Assays
Cat. No. / ID: 337812
Laboratory-verified qPCR assays for measuring changes in copy number
ログイン アカウントの価格を確認するには
qBiomarker Copy Number PCR Assaysは分子生物学的アプリケーション用であり、疾病の診断、予防、あるいは治療に使用することはできません。
GeneGlobeで探索・設定する
適切なターゲット特異的アッセイおよびパネルを探すか、またはターゲットをカスタムデザインし、お客様のご興味のある生物学的ターゲット評価にお使いください。
特徴
- ゲノム全領域をターゲットする1000万以上のアッセイが入手可能
- ウエットベンチによる試験済みアッセイ
- 簡単なリアルタイムPCR手法
- ウェブベースの無料のデータ解析ソフトウェア
製品詳細
qBiomarker Copy Number PCR Assaysは、個別の目的遺伝子(GOI)または目的領域(ROI)において特異的で、正確かつ再現性があり、解析しやすいコピー数変化結果を獲得する方法を提供します。すべてのアッセイは実験室で検証済みで、マイクロアレイのフォローアップ研究、特定ターゲットのスクリーニング、および関連研究ですぐに使用できます。
パフォーマンス
qBiomarker Copy Number PCR Assaysはすべて、リアルタイムPCR結果の正確さに影響する次のようないくつかの特性についてウエットベンチで試験済みです:特異性、広いダイナミック・レンジ、均一で高い増幅効率。アッセイ品質のラボ検証により、qBiomarker Copy Number PCR Assaysで信頼性の高い結果が確実に得られます。
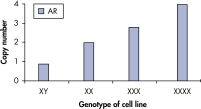
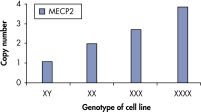
qBiomarker Copy Number PCR Assaysは、すでに細胞遺伝学的方法によって同定された染色体異常を含む細胞株において、異数性を正確に同定しました。X染色体上に共に存在する AR と MECP2のアッセイは、1、3、4コピーのX染色体を持つ細胞株での遺伝子コピー数を正確に定量しました。
図参照
原理
すべてのqBiomarker Copy Number PCR Assayは、独自のゲノム領域内に設定されています。qBiomarker Multicopy Reference Copy Number PCR Assay(MRef、別途購入可能)は、DNAインプットの卓越した正規化を実現し、qBiomarker Copy Number PCR Assaysと併用することを推奨します。MRefアッセイは、ヒトゲノムに40回以上存在する安定した配列を認識し、そのコピー数は局所的なゲノム変化の影響を全く、あるいは殆ど受けません。テスト中にこのリファレンスアッセイを同時に実施することにより、ΔΔCT法を用いて、特定のターゲットに対してコピー数のコールあるいは相対的なコピー数変化のコールを正確に行なえます。オールインワンタイプで即使用可能なqBiomarker SYBR® Green PCR Mastermixesは、最小限のプライマーダイマーで遺伝子座特異的な最大増幅を提供するために最適化されています。
操作手順
新鮮、凍結、FFPEサンプルからのゲノムDNAを単離します。各サンプルの溶液を、適切な即使用可能なqBiomarker SYBR Green MastermixおよびqBiomarker Copy Number PCR Assayと混和します。別のチューブに、一定量の各サンプルとマスターミックスならびにqBiomarker Multicopy Reference Copy Number PCR Assayを混和します。リアルタイムサイクラーを実行し、無料のデータ解析ツールを用いて、テストサンプルとリファレンスゲノムを比較するΔΔCT法により、コピー数の変化を計算します。
アプリケーション
qBiomarker Copy Number PCR Assaysは、新鮮、凍結、FFPEサンプルから、個別の遺伝子座におけるコピー数異常や多型の正確な検出に最適です。
裏付けデータと数値
qBiomarker Copy Number PCR Assays accurately identify aneuploidy.
qBiomarker Copy Number PCR Assays accurately identify aneuploidy.
リソース
MSDS (1)
ホワイトペーパー (1)
キットハンドブック (1)
機器テクニカル資料 (1)
Safety Data Sheets (1)
Certificates of Analysis (1)
Instrument Technical Documents (1)
Kit Handbooks (1)
White Papers (1)
FAQ
What sample types can I test on qBiomarker Copy Number PCR Arrays?
Will qBiomarker Copy Number Assays work with heterogeneous samples like mixtures of tumor and normal tissue?
Can I use genomic DNA from fixed samples with qBiomarker Copy Number PCR Arrays?
Why do qBiomarker Copy Number PCR Arrays have assays in quadruplicate?
What qPCR mastermix should I use with the qBiomarker Copy Number PCR Arrays and Assays
Can I use DNA isolated from an AllPrep DNA/RNA Kit with qBiomarker Copy Number PCR Arrays and Assays
How should I analyze the data generated from a qBiomarker Copy Number PCR Array experiment?
Why is the copy number call lower than expected?
What could have gone wrong if the CT values are unusually high for all wells in a sample?
What testing should be performed in order to assess the quality of a DNA sample?
How much DNA should I use in a qBiomarker Copy Number PCR Assay?
Can I use amplified genomic DNA with qBiomarker Copy Number PCR Arrays?
How do you choose which assays to include on the arrays?