QuantiTect Reverse Transcription Kit
遺伝子発現解析のための高感度な2ステップリアルタイムRT-PCR用cDNAの迅速合成
遺伝子発現解析のための高感度な2ステップリアルタイムRT-PCR用cDNAの迅速合成
Cat. No. / ID: 205311
QuantiTect Reverse Transcription KitはゲノムDNA除去を組み込んだcDNA合成を迅速かつ簡便な方法で実現します。RNAサンプル中のゲノムDNAコンタミはgDNA Wipeout Bufferで効率的に除去されます。Quantiscript Reverse Transcriptase、Quantiscript RT Buffer、画期的なRT Primer Mixにより、迅速かつ効率的な逆転写反応に必要な全ての成分が提供されます。合成されたcDNAはリアルタイムPCRでの使用に至適化され、mRNA転写物のどの領域の標的も確実に定量できます。
QuantiTect Reverse Transcription Kitを用いることにより、RNAサンプル中に混入しているゲノムDNAはユニークなgDNA Wipeout Bufferで効率的かつ迅速に除去されます(図" 効果的なゲノムDNA除去による正確なリアルタイムRT-PCR")。ゲノムDNAの排除は正確な遺伝子発現結果を得るために極めて重要であり、RNA特異的プライマーまたはプローブのデザインは必ずしも可能とは限りません。gDNA Wipeout Bufferを用いると、RNAサンプルの精製中あるいは精製後に別途にDNase分解を行なう必要がないため、時間および経費を節約できます。
Quantiscript RT Bufferと組み合わせて用いることにより、Quantiscript Reverse Transcriptaseの高いRNAアフィニティーはどのようなRNAテンプレートからも高い収量でcDNAを合成できます(表"より少量の転写物からより高いcDNA収量")。GC含有率が高いあるいは複雑な二次構造を持つ逆転写が困難なテンプレートでさえも問題なく逆転写できます。
| IL12AのCT値 (低発現量) | IL1RNのCT値 (高発現量) | |||
| スタートRNA(ng) | QIAGEN | Supplier AII | QIAGEN | Supplier AII |
|---|---|---|---|---|
| 1000 | 30.9 | 32.0 | 23.1 | 24.9 |
| 100 | 34.2 | 35.4 | 26.3 | 26.6 |
| 10 | 37.8 | 46.8 | 29.7 | 30.3 |
| 1 | 非検出 | 非検出 | 32.4 | 34.5 |
RT Primer Mixには全てのRNA転写物の5'領域も含むあらゆる領域からcDNA合成を実現する特別に至適化されたoligo-dTオリゴ-dTとランダムプライマーのミックスが含まれています(図" 12.5 kbの転写物の5'領域における標的を高感度で検出")。他社のキットと比較して、QuantiTect Reverse Transcription Kitは増幅する標的領域が転写物のどこに局在しているかに関係なく、リアルタイムPCR解析用のcDNAテンプレートを高い収量で合成し、発現量が少ない遺伝子の検出感度を上昇させます(図" より高い感度の2ステップリアルタイムRT-PCR")。また、QuantiTect Reverse Transcription Kitは、再現性のより高いリアルタイムRT-PCRを実現します。
QuantiTect Reverse TranscriptaseはOmniscriptとSensiscript Reverse Transcriptaseのブレンドで、RNAに対して高いアフィニティーを有し、幅広いRNA量(10 pg~1 µg)からcDNA合成が可能です。他社のキットと比較して、QuantiTect Reverse Transcription Kitは増幅する標的領域が転写物のどこに局在しているかに関係なく、リアルタイムPCR解析用のcDNAテンプレートを高い収量で合成します。GC含有率が高いあるいは複雑な二次構造を持つ逆転写が困難なテンプレートでさえも問題なく逆転写できます。また、QuantiTect RT BufferはリアルタイムPCRバッファーと互換性があるようにも至適化されています。
リアルタイムRT-PCRによる遺伝子発現アッセイで正確な結果を得るためには、cDNAのみを増幅し検出することが重要です。ゲノムDNAによるアッセイへの悪影響はエクソン/エクソン境界にまたがるプライマーやプローブをデザインすることにより避けることができます。しかし、この方法が不可能な場合もあり(例えばシングルエクソン遺伝子からのcDNA)、スタートとなるRNAサンプルがゲノムDNAを含まないことが必須です。QuantiTect Reverse Transcription Kitを用いることにより、RNAサンプル中に混入しているゲノムDNAはユニークなgDNA Wipeout Bufferで効率的かつ迅速に除去されます。RNAサンプルの精製中あるいは精製後に別途にDNase分解を行なう必要がないため、時間および経費を節約できます。RNA特異的なプライマーやプローブのデザインも不要です。
| 構成 | 利点 |
|---|---|
| gDNA Wipeout Buffer | リアルタイムRT-PCRのみでRNAを検出 |
| Quantiscript Reverse Transcriptase | 幅広いRNA量(10 pg~1 µg RNA)の使用 高感度 |
| Quantiscript RT Buffer | 増幅困難なテンプレートの読み取り |
| RT Primer Mix | 転写物の全ての領域、5'領域からもcDNAを合成 |
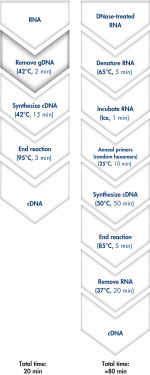
QuantiTect Reverse Transcription Kitを用いれば、ゲノムDNA除去とcDNA合成はわずか20分で完了します(フローチャート" 迅速で簡便なcDNA合成")。両反応とも同一のインキュベーション温度で行なうことができ、マスターミックスによるセットアップができるため、操作は迅速かつ簡便です。
QuantiTect Reverse Transcription Kitには、迅速なcDNA合成に必要なものが全て含まれています。精製したRNAはgDNA Wipeout Bufferで短時間インキュベートし、混入したゲノムDNAを効率的に除去します。他の方法と比較し、逆転写反応において、Quantiscript Reverse Transcriptase、Quantiscript RT Buffer、RT Primer Mixで調製したマスターミックスを用い、RNAサンプルを直接使用します。Quantiscript Reverse Transcriptaseを用いると、複雑な2°構造でもRNAは低温で転写され、RNAは分解されません。反応全体は42℃で起こり、95℃で不活化されます。RNA変性、プライマーアニーリング、RNase H分解の追加ステップは必要ありません。
QuantiTect Reverse Transcription Kitはレーザーマイクロダイセクションサンプルや組織生検を含む全ての種類のスタートサンプルから感度および効率の良いリアルタイムRT-PCRを実現します。
Genomic DNA removal and cDNA synthesis take only 20 minutes with the QuantiTect Reverse Transcription Kit. The procedure is fast and convenient since both reactions are run using the same incubation temperature and are set up using master mixes. In contrast, the procedure for the kit from Supplier I is much longer and requires more "hands-on time" due to additional pipetting steps and frequent changes in incubation temperature.
| Features | Specifications |
|---|---|
| Applications | Quantification of (even low-abundance) transcripts |
| Sample/target type | RNA template |
| Enzyme activity | Reverse transcription |
| Real-time or endpoint | Real time |
| Reaction type | Two-step, cDNA production, genomic DNA digestion |
| Single or multiplex | Single |
| Mastermix | No |