✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
特徴
- 最小限の溶出量
- 迅速な操作と容易な取り扱い
- 再現性のある高い回収率
- ゲル解析をより簡便化するGelPilot Loading Dye付き
製品詳細
MinElute PCR Purification Kitは、スピンカラム、バッファー、コレクションチューブで構成され、シリカメンブレンによりPCR 産物(70 bp~4 kb)の精製を実現します。スピンカラムは微量(わずか10 µl)で溶出できるようにデザインされているので、高濃度のDNAが高収量で得られます。オプションのpH指示薬により、DNAがスピンカラムに結合する最適なpHを簡単にチェックできます。本キットはQIAcubeで完全自動化が可能です。
パフォーマンス
MinElute PCR Purification Kitには、PCR生成物クリーンアップ用のスピンカラムが含まれています。マイクロ遠心機または吸引マニホールドを使用することにより、高濃度なDNAフラグメント(70 bp~4 kb)が迅速に得られます(4 kb以上のDNAフラグメントの精製には QIAquick PCR Purification Kit をご利用ください)。
図参照
原理
MinElute PCR Purification Kitでは、高塩濃度バッファーによりDNAを結合し、低塩濃度バッファーあるいは水によりDNAを溶出するシリカゲルメンブレンを利用しています。シリカメンブレンテクノロジーにより樹脂漏れ、および懸濁液関連の問題や不便さが解消されます。
Gel Loading Dye
より迅速で簡便なサンプル処理と解析を実現するため、ゲル電気泳動用のローディング色素が付いています。GelPilot Loading Dye は3 種類のマーカー色素(xylene cyanol、bromophenol blueおよびorange G)が入っており、短いDNAフラグメントのゲルからの流出を回避でき、アガロースゲルの泳動時間の至適化が簡単に行なえます(図 " GelPilot Loading Dye")。
図参照
操作手順
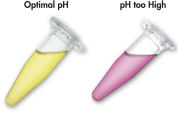
MinEluteシステムでは結合-洗浄-溶出という簡単な操作だけでDNAのクリーンアップが可能です。結合バッファーをPCRサンプルあるいは他の酵素反応液に直接添加し、混合液をMinElute Spin Column にアプライします。ゲル切片をpH指示薬を含んだバッファーに溶解させると、DNA 結合の至適pH を容易に決めることができます (図 “pH指示薬”)。DNAは添付のバッファー中の高塩濃度の条件下でシリカゲルメンブレンに吸着します。不純物は洗い流され、純粋なDNAを添付の低塩濃度溶出バッファー、あるいは水で溶出します。 得られたDNAは、その後のアプリケーションに即使用可能です。
操作法
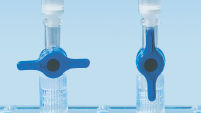
MinElute Spin Column は2つの簡便な操作法オプションのためにデザインされました(フローチャート"MinElute操作手順")。スピンカラムは簡便な卓上型マイクロ遠心機あるいはQIAvac 24 PlusやQIAvac Luer Adapter付きQIAvac 6S (図 "Spin Column操作オプション A、 B、 C、 D、および E")のようなルアーコネクター付き吸引装置で使用でき、QIAcubeでの完全自動化も可能です。
図参照
アプリケーション
MinEluteシステムで精製したDNAフラグメントは以下のようなアプリケーションに即使用可能です。
- 次世代シークエンシングを含むシークエンシング
- マイクロアレイ解析
- ライゲーションやトランスフォーメーション
- 制限酵素分解
- 標識反応
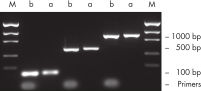
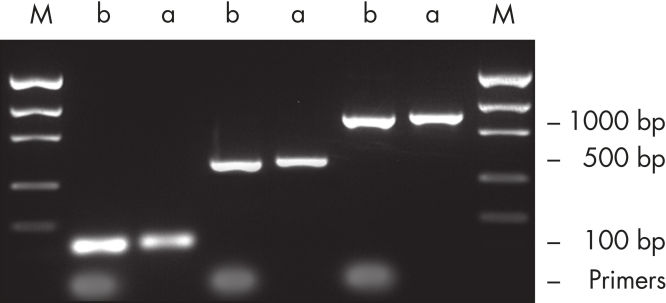
裏付けデータと数値
効率的なプライマー除去。
Specifications
| Features | Specifications |
|---|---|
| Binding capacity | 5 µg |
| Sample type: applications | DNA、オリゴヌクレオチド:PCR反応 |
| Elution volume | 10 µl |
| Fragment size | 70 bp ~ 4 kb |
| Recovery: oligonucleotides dsDNA | 回収:オリゴヌクレオチド、dsDNA |
| Format | チューブ |
| Technology | シリカテクノロジー |
| Processing | Manual |
| Removal <10mers 17–40mers dye terminator proteins | 除去<40mers |