HotStarTaq DNA Polymerase (250 U)
Cat. No. / ID: 203203
特徴
- 最小限の至適化
- 高いPCR特異性
- 取り扱いが簡単、室温でのセットアップが可能
製品詳細
HotStarTaq DNA Polymerase は抗体によるホットスタートシステムではなく、化学修飾によるホットスタートを利用しているため、最初の熱活性化ステップまではポリメラーゼ活性は全くありません。HotStarTaq DNA Polymerase には、非特異的な増幅産物、プライマーダイマー、バックグラウンドを最小限に抑える画期的なQIAGEN PCR Buffer が入っています。また増幅困難なテンプレート(例;GC リッチ)の効率的な増幅を実現するQ-Solutionも含まれています。
パフォーマンス
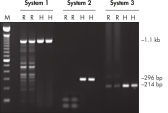
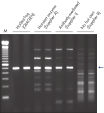
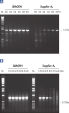
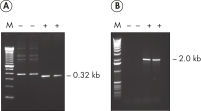
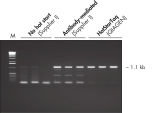
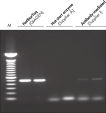
HotStarTaq DNA Polymeraseは全てのロットで、低コピー数のターゲットを増幅する実験によりPCR特異性の厳密さや再現性をチェックするなど、広範囲の品質管理テストを受けています。HotStarTaq DNA Polymeraseは他社のキットに比べて高性能で、特異性と感度の高いホットスタートPCRを実現します (図“ 異なるプライマー/テンプレートシステムにおける高特異性”、“ 卓越した性能”および表)。キットに付属の画期的なPCRバッファーは、様々なPCR条件において特異性を実現し、至適化は最小限に抑えられます(図 “ 幅広い至適アニーリング温度” および“ 異なったマグネシウム濃度への適応”)。また、キットに付属のQ - Solutionにより、不適切なPCR条件を改善することができます(図 “ 増幅困難なテンプレートの増幅”)。これらの成分により様々なアプリケーションで特異的な増幅を実現します(図 “ RT-PCRにおけるホットスタートの効果”および “ 高感度のシングルセルPCR”)。
| HotStarTaq DNA Polymerase | ホットスタート酵素AII社 | 抗体利用 | マニュアル | ワックスバリア | |
|---|---|---|---|---|---|
| 特異的な増幅 | ++ | + | + | +/– | +/– |
| PCR至適化の不要性 | ++ | +/– | +/– | – | – |
| 取り扱いの簡便さ | ++ | ++ | + | – | – |
HotStarTaq DNA Polymerase 詳細データ
濃度:5 units/µl
組み換え酵素:Yes
基質アナログ:dNTP、ddNTP、dUTP、biotin-11-dUTP、DIG-11-dUTP、 fluorescent-dNTP/ddNTP
エクステンション速度:72℃ で2~4 kb/min
半減期:97℃で10分、94℃で60分
増幅効率:≥105 倍
5'–>3' エキソヌクレアーゼ活性:Yes
余分なA付加:Yes
3'–>5' エキソヌクレアーゼ活性:No
ヌクレアーゼの混入:No
RNasesの混入:No
プロテアーゼの混入:No
自己プライマー活性:No
図参照
原理
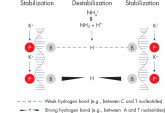
Taq DNA Polymeraseを修飾したHotStarTaq DNA Polymeraseは、ホットスタートPCR において高い特異性を実現します。本キットには、2種類の陽イオンを含む画期的なPCRバッファー、Q-Solution、MgCl2が入っています。
HotStarTaq DNA Polymerase
HotStarTaq DNA Polymeraseは、常温では不活性状態でポリメラーゼ活性はありません。この特性により、PCR セットアップや最初のPCR サイクル中の低温での非特異的なプライマーのアニーリングやプライマーダイマーの形成を回避できます(図“ ホットスタートPCRで最高のパフォーマンスを実現”および“ 異なるプライマー/テンプレートシステムにおける高特異性”)。HotStarTaq DNA Polymeraseは、95℃、5 分間のインキュベーションステップで活性化され、このステップは既存のサーマルサイクリングのプログラムに容易に導入できます。
QIAGEN PCR Buffer
QIAGEN PCR Buffer は、各PCRサイクルでアニーリングステップ中にプライマー特異的な結合の割合を非特異的な結合に対して高めることにより、各PCRサイクルの特異的な増幅を維持します(図“ プライマーアニーリングの特異性増大”)。このバッファーはユニークな配合比のKClと (NH4)2SO4を含み、従来のPCRバッファーに比べ、幅広いアニーリング温度やMg2+濃度の範囲で厳密で特異的なプライマーアニーリングを実現します。従って、異なるアニーリング温度あるいはMg2+ 濃度を用いて行なうPCRの至適化は最小限ですみ、または不要なこともあります(図“ 幅広い至適アニーリング温度”および“ 異なったマグネシウム濃度への適応”)。
Q-Solution
HotStarTaq DNA Polymeraseに入っているQ-Solutionは、DNAの融解を変更することにより増幅困難なテンプレートの増幅を促進する革新的なPCR添加物です。このユニークな試薬により、高度な二次構造をもつテンプレートやGC リッチなテンプレートなどにより生じる不適切なPCR 条件が改善されることがあります(図“ 増幅困難なテンプレートの増幅”)。DMSOのような汎用されているPCR添加物と異なり、Q-Solutionは一定の濃度で作用し、毒性もなく、PCR純度は保証されています。Q-Solution の添加により、PCR のフィデリティは損なわれません。
図参照
操作手順
図参照
アプリケーション
HotStarTaq DNA Polymeraseは、増幅などの高度なアプリケーションを含む様々なアプリケーションに最適です:
- 複雑なゲノムテンプレート
- 複雑なcDNAテンプレート(例;RT-PCR)
- 低コピーのターゲット(例;シングルセルPCR)
- マルチプレックスPCR
裏付けデータと数値
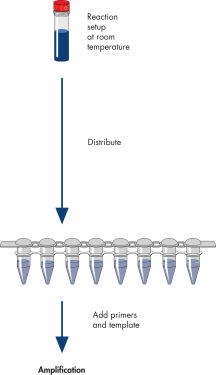
HotStarTaq procedure.
Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, RT-PCR, Complex genomic templates, very low-copy targets |
| With/without hotstart | With hotstart |
| Reaction type | PCR amplification |
| Sample/target type | Genomic DNA and cDNA |
| Real-time or endpoint | Endpoint |
| Enzyme activity | 5' -> 3' exonuclease activity |
| Mastermix | No |
| Single or multiplex | Single |