dPCR LNA Mutation Assay (200)
Cat. No. / ID: 250200
Features
- Duplex assay design detects mutated and wild-type sequences
- Choice of FAM + HEX or Atto 550 + ROX fluorescent dye combinations
- LNA-enhanced primers and probes increase assay specificity and sensitivity
- Wet-lab tested dPCR assays available for more than 200 targets
- Suitable for use with circulating tumor DNA, liquid biopsy, FFPE and other tissue samples
- Intended for use with QIAcuity MasterMix
Product Details
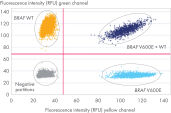
dPCR LNA Mutation Assays enable detection of individual sequence mutations selected from comprehensive curated databases such as COSMIC. The assay designs have been dPCR wet-lab tested, and the resulting data is available. The sensitivity for detecting the target mutation is 0.1% in a wild-type background in one well of a QIAcuity Nanoplate. Even higher sensitivity can be achieved by splitting the reaction into multiple wells and combining the analysis. The choice of two different fluorescent dye combinations allows detection of mutant and wild-type sequences as well as multiplexing analysis of two target mutations in one well.
The assays are available in tube format as a ready-to-use, 30x-concentrated assay, consisting of two primers and probes for mutant and wild-type. dPCR LNA Mutation Assays are to be used with the QIAcuity MasterMix.
Performance
dPCR allows for increased sensitivity in the detection and absolute quantification of rare events/sequence variants, making it possible to detect a mutation presence at 0.1% in a background of wild-type genomic DNA. The partitioning enables an increase of the specific concentration of the target. The end-point amplification and detection over noise, with a binary positive/negative target calling allows precise quantification, which is independent from amplification performance (cause for example by PCR inhibitors).
Principle
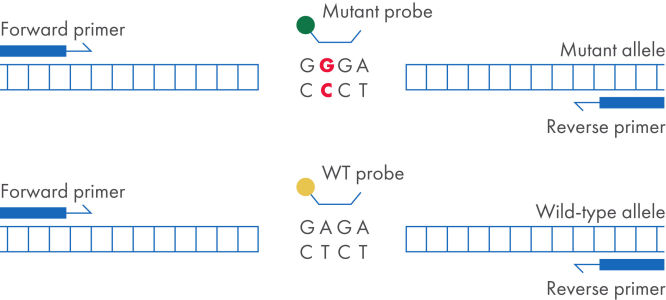
The dPCR LNA Mutation Assays are duplex, hydrolysis probe-based assays that identify the presence of specific mutation sequences. One probe detects the mutant allele, and the other probe detects the wild-type allele. Digital PCR provides the most sensitive and reliable method for detecting a mutant DNA sequence in a wild-type background. dPCR LNA Mutation Assays use LNA-enhanced primers and probes, which increase assay specificity and sensitivity, making it possible to detect a mutation present at 0.1% in a background of wild-type genomic DNA in a single nanoplate well.
Two fluorescence detection options enable multiplex analysis of two mutation targets in one well. During product configuration, the choice of FAM + HEX or Atto 550 + ROX for detecting mutant + wild-type is available.
Procedure
To set up the dPCR experiment, add an aliquot of the genomic DNA sample (isolated from fresh, frozen or fixed samples) to the QIAcuity MasterMix and the dPCR LNA Mutation PCR Assay. Each reaction is analyzed in one or multiple wells of a QIAcuity Nanoplate.
After loading and sealing the nanoplate, select the recommended cycling program on the QIAcuity instrument. In the absolute quantification analysis type of the QIAcuity Software Suite, the copies per microliter result is shown. The calculated fractional abundance of the mutant target and corresponding data figures are available in the analysis type mutation analysis.
Applications
dPCR LNA Mutation PCR Assays are highly suited for rapidly and accurately identifying individual specific sequence mutations present in DNA from fresh, frozen or fixed samples.
Supporting data and figures
dPCR LNA Mutation Assay setup.