EZ1&2 DNA FFPE Kit (48)
Cat. No. / ID: 954404
Features
- High recovery of amplifiable DNA
- Paraffin removal without xylene or similar solvents
- Option to remove uracil (deaminated cytosine) artifacts
- Ready-to-use DNA for PCR, digital PCR (dPCR) and next-generation sequencing (NGS)
- Automated heating and reagent-pipetting pretreatment steps, available on the EZ2 Connect
Product Details
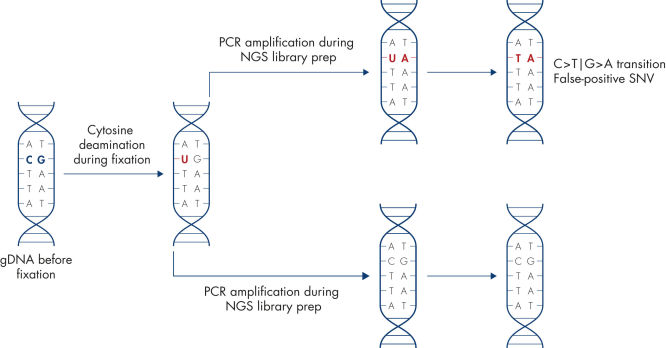
Purify large amounts of amplifiable DNA from hard-to-lyse formalin-fixed, paraffin-embedded (FFPE) tissue. The EZ1&2 DNA FFPE protocol uses double lysis to recover DNA effectively, while the optional uracil-N-glycosylase (UNG) step removes deaminated cytosine artifacts to limit the risk of nucleotide read errors.
The EZ1&2 DNA FFPE protocols are automatable on either the EZ1 Advanced XL or the EZ2 Connect.
The EZ2 Connect features cardless protocols, expanded applications and remote connectivity options. Click here to learn more about the EZ2 Connect, or contact your sales representative to discover how easily you can get an EZ2 Connect for your lab.
Performance
Under UV-vis, fluorometric, and qPCR analysis, most of the samples processed with the EZ1&2 DNA FFPE Kit showed a higher abundance of dsDNA (see figure “ Optimized for PCR performance”).
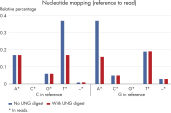
For NGS analysis, the EZ1&2 DNA FFPE UNG Kit significantly reduces artificial C→T/G→A transitions, which commonly occur in FFPE material due to cytosine deamination (see “ Artificial C→T/G→A transitions”). This helps prevent false-positive reports of single-nucleotide variants (SNVs) in NGS (see figure “ Reduction in artifactual C→T | G→A transitions”).
See figures
Principle
The EZ1&2 DNA FFPE Kit maximizes DNA yields from limited sample inputs in two ways: By implementing a two-step lysis procedure, it helps ensure high DNA extraction even from difficult-to-lyse samples. In addition, its wash-free no-solvent deparaffinization technique minimizes the risk of losing of any sample material, which is difficult to avoid with multiple-wash solvent-based deparaffinization methods.
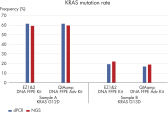
Crosslink removal further increases the amount of recovered amplifiable DNA (see figure “ Optimized for PCR performance”). In addition, it also makes the recovered DNA especially suitable for NGS analysis (see figure “ Reliable dPCR and NGS results”).
See figures
Procedure
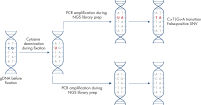
The EZ1&2 DNA FFPE Kit fully automates the bind, wash and elute steps on the EZ1 Advanced XL or the EZ2 Connect. The preparation steps (i.e., Proteinase K digestions, crosslink removal and RNase A digestion) are done manually on the EZ1 Advanced XL; on the EZ2 Connect, many of these preparation steps can be automated (see figure “ EZ1&2 DNA FFPE workflow”).
See figures
Applications
DNA isolated with the EZ1&2 DNA FFPE Kit can be used immediately for PCR, dPCR or NGS. Alternatively, it can be stored at −30 to −15°C.
Supporting data and figures
Artificial C→T/G→A transitions.
Sample fixation can lead to artifacts in the DNA sequence. The most common artifact in FFPE tissues is the deamination of cytosine to uracil, which results in a single nucleotide variation (SNV).
Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, digital PCR, next-generation sequencing |
| Elution volume | 60 and 100 µl |
| Format | Magnetic bead |
| Main sample type | Formalin-fixed paraffin-embedded tissue samples |
| Processing | Automated with the EZ1 Advance XL or EZ2 Connect |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein | Genomic DNA |
| Technology | Silica technology |
| Sample amount | Tissue sections, each with a thickness of 5–10 µm, for a total volume of 4 mm3 |