Products
Features
- Over 1.3 million assays provide the broadest and best coverage of human, mouse and rat mRNA and lncRNA transcripts in Ensembl
- Exceptional sensitivity and specificity using short, LNA-enhanced primers
- Accurate detection over a wide dynamic range starting at 1 RNA copy
- Optimized to eliminate non-specific amplification
- Robust and reproducible results in 2 hours using the QuantiNova SYBR® Green PCR reagents
Product Details
QuantiNova LNA PCR Assays are setting new standards in SYBR® Green-based qPCR gene expression analysis. Choose from the broadest selection of predesigned assays for accurate and sensitive detection of any human, mouse or rat mRNA or lncRNA – from general transcript detection to differentiation of specific transcript isoforms. LNA enhancement allows us to design shorter primers, which are more flexible to position on the target. As a result, we can optimize our designs for specific transcript detection or target differentiation. Plus, thorough design validation ensures optimal performance and robust detection, and assays for all popular mRNAs and lncRNAs undergo wet-lab validation. Optimized using the QuantiNova RT and SYBR® Green reagents, the simple QuantiNova LNA PCR workflow can be completed in just 2 hours and is robust enough to support room-temperature setup, automation and delayed run of assays.
Are you planning to use the QuantiNova LNA PCR Assays for digital PCR analysis? You can find dPCR-specific product and performance details on the dedicated dPCR catalog page.
Need a quote for your research project or would you like to discuss your project with our specialist team? Just contact us!
Performance
LNA enhances qPCR performance
QuantiNova LNA PCR Assays have been developed using stringent design criteria and lab-validated algorithms. The rigorous assay selection process ensures that each primer set delivers the highest specificity and efficiency for the most reliable and accurate results. Tm-normalization and LNA enhancement give the primers a higher binding affinity than standard DNA primers, dramatically increasing assay sensitivity and specificity.
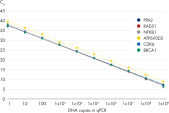
This increased sensitivity ensures excellent amplification efficiencies down to 1 RNA molecule, making it easier for you to detect low-abundance targets such as lncRNAs from less starting material (see figure QuantiNova LNA PCR Assays provide accurate, sensitive and linear quantification of targets over a wide dynamic range and figure QuantiNova LNA PCR Assays enable both high-expression and low-expression detection of mRNA). Increased specificity from the clever placement of LNA gives you a high signal-to-noise ratio, allowing discrimination of sequences that differ by only a single nucleotide and eliminating non-specific amplification and primer-dimer formation.
The high binding affinity of LNA bases increases the flexibility of primer placement on the transcript, so we are able to use intelligent positioning to design assays for otherwise difficult-to-analyze targets. This gives you better target discrimination and reliable quantification, even for AU-rich targets, low-abundance transcripts, targets with high secondary structure and highly complex samples.
Optimized with QuantiNova SYBR® Green PCR reagents
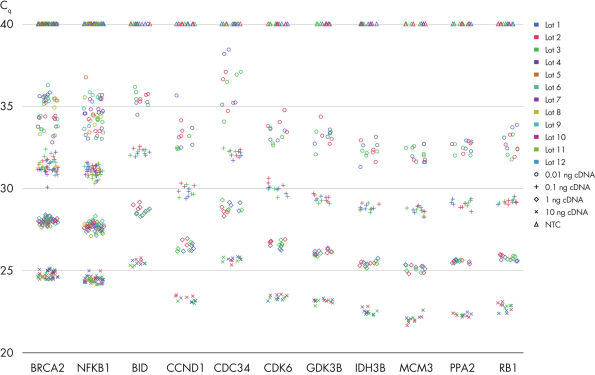
QuantiNova LNA PCR Assays were developed to give optimal results and the highest reproducibility when used with the QuantiNova SYBR® Green PCR reagents (see figure Demonstrated lot-to-lot reproducibility). For 1-step qRT-PCR, this includes the QuantiNova SYBR® Green RT-PCR Kit. For 2-step qRT-PCR, this includes the QuantiNova Reverse Transcription Kits and QuantiNova SYBR® Green PCR Kits.
The QuantiNova hot-start mechanism enables successful qPCR, even for difficult templates, such as those with high GC-content or complex secondary structure – ideal for lncRNA analysis (see figure Consistently high efficiencies throughout the expected GC content range of the human transcriptome). The hot start also allows reaction setup at room temperature, and is robust enough to support automated setup and delayed run of assay plates. Plus, the built-in visual pipetting color indicator helps prevent human pipetting errors.
Trust the LNA design experts
Twenty years of LNA design experience has enabled us to develop and optimize our sophisticated LNA design algorithm, which incorporates over 50 different parameters – all thoroughly lab-validated against highly stringent performance criteria – to guarantee the most optimal assay for successful target detection. We have selected up to three different assay designs for each transcript and fully wet-lab validated all popular mRNA and lncRNA target assays. QuantiNova LNA PCR Assays simply work, with no need to spend your time or money on optimization.
Fast and easy workflow
The fast and easy 2-hour workflow with minimal hands-on time saves time and labor and is suitable for automation. The simple protocol has been tested for compatibility on all major brand qPCR cyclers (see figure QuantiNova LNA PCR Assays deliver consistent performance across all thermal cycler instruments). Furthermore, the cDNA generated in the RT step can be used across the entire QuantiNova LNA PCR System, allowing seamless transition from assays to panels depending on your research needs, saving you time and sample.
See figures
 QuantiNova LNA Probe PCR Assays enable both high-expression and low-expression detection of mRNA.
QuantiNova LNA Probe PCR Assays enable both high-expression and low-expression detection of mRNA.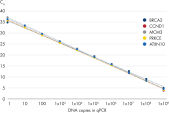 Demonstrated lot-to-lot reproducibility.
Demonstrated lot-to-lot reproducibility. Consistently high efficiencies throughout the expected GC content range of the human transcriptome.
Consistently high efficiencies throughout the expected GC content range of the human transcriptome.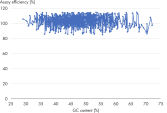 QuantiNova LNA PCR Assays deliver consistent performance across all thermal cycler instruments.
QuantiNova LNA PCR Assays deliver consistent performance across all thermal cycler instruments.
Principle
High-performance SYBR® Green-based detection
QuantiNova LNA PCR Assays use SYBR® Green-based detection, which, when used with a highly specific PCR system, well-designed primers and optimized reaction conditions, offers several advantages over hydrolysis probe detection. First, it is possible to perform melting curve analysis immediately following the PCR, so you can confirm high specificity of the reaction and rule out formation of primer–dimers or non-specific amplicons. It also provides higher flexibility in the assay design, because you only need two primers, instead of two primers and a probe. Furthermore, while the primers ensure the specificity of the detection, the dye’s binding properties enable analysis of many different targets without having to synthesize target-specific, labeled probes, reducing your assay setup and running costs.
Most comprehensive and specific coverage
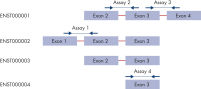
Our proprietary algorithm has been used to design over 1.3 million QuantiNova LNA PCR Assays to provide the most sensitive, accurate and effective mRNA and lncRNA analysis. The predesigned assays cover most transcripts in the Ensembl database for human, mouse and rat genes, enabling PCR-based gene expression studies in the greatest depth possible. Most of the assays are intron-spanning when possible and detect only RNA. Assays that do not span an intron are designated as such, and if there is one exon in the target, unwanted signals can be easily eliminated using the QuantiNova Reverse Transcription Kit with the integrated gDNA removal step.
Choosing the right assay for your target
Predesigned QuantiNova LNA PCR Assays let you accurately and sensitively detect any human, mouse or rat mRNA or lncRNA, no matter what level of analysis you need: general transcript detection, detection or a specific transcript or differentiation of transcript isoforms.
Most human, mouse and rat genes have only one transcript, but for those with multiple transcripts, we selected up to three assays per transcript using the same algorithm. The design and primer positioning of these assays differ to match various usage requirements. Our assay selection guide helps you quickly identify the best assay from each category. After searching for assays for your target, simply review the results and look for the recommended assay that matches your intended use:
- Best coverage: for general transcript detection
- Best transcript assay: for detection of a specific transcript
- Best transcript-specific assay: for differentiation between specific transcript isoforms
Refer to the table below for more details.
| Gene-covering assays | Transcript-specific assays | ||
|---|---|---|---|
| QIAGEN’s recommended assay | Marked as “Best coverage” | Marked as “Best transcript assay” | Marked as “Best transcript-specific assay” |
| Description of assay design and coverage | Covers most of the biologically relevant transcripts of the given gene | Highly optimized to specifically detect the transcript of interest, targeting more isoforms of the transcript | Splice variant transcript-specific |
| When to choose | For general transcript screening and determining whether the gene of interest is expressed in a sample; for detection of as many transcripts and isoforms of a gene as possible | For detection of a specific transcript | For differentiation between specific isoforms of a transcript |
Complimentary data analysis
The complimentary, web-based data analysis tool in the GeneGlobe Data Analysis Center includes a user-friendly wizard to guide you step-by-step through the normalization and analysis of your data and generates publication-ready results figures.
Reference Gene Assays for any study
A wide selection of human, mouse and rat Reference Gene Assays are available to enable high-quality data normalization and ensure reliable results. These assays target endogenous coding RNAs, long non-coding RNA and small nucleolar RNA molecules that are typically constitutively expressed in a wide variety of tissues. The Reference Gene Assays are FAM labeled and have been functionally validated as reference genes for the QuantiNova LNA PCR system and work optimally with the QuantiNova RT and PCR reagents.
Normalization of mRNA/lncRNA qPCR results
Normalization removes technical and biological inter-sample variation unrelated to the biological changes under investigation. Proper normalization is critical for correct analysis and interpretation of results from real-time PCR experiments. Most commonly, stably expressed reference genes are used for normalization.
It is generally recommended to test several endogenous control reference gene candidates before setting up your actual mRNA/lncRNA expression analysis. These candidates should be chosen from genes expected to be stably expressed over the whole range of samples under investigation. They could be stably expressed mRNAs or lncRNAs selected based on literature or preexisting data (e.g., NGS or qPCR panel screening). The QuantiNova LNA PCR system offers validated reference gene assays for RNAs that tend to be stably expressed and are therefore good candidates as reference genes.
All reference gene candidates should be empirically validated for each study. One option for normalizing PCR panel when profiling a large number of mRNAs/lncRNAs is to normalize against the global mean – the average of all expressed mRNAs/lncRNAs. This can be a good option in samples with a high call rate (expressed genes) but should be used with caution in samples with low call rates. It is also not a good option in samples for which the general gene expression level is changed. Further guidance on normalization can also be found in the GeneGlobe Data Analysis Center.
Procedure
Choose one- or two-step qRT-PCR
QuantiNova LNA PCR Assays have been optimized using the time-tested and high-performance QuantiNova chemistry. Best results are obtained when reverse transcription is performed with the QuantiNova Reverse Transcription Kit. The resulting cDNA is then quantified by qPCR using the master mixes of the QuantiNova SYBR® Green PCR Kit combined with your choice of QuantiNova LNA PCR Assay.
For 1-step qRT-PCR, we recommend using the QuantiNova SYBR® Green RT-PCR Kit, which requires no further optimization of reaction conditions. Simply add primers, RNA template and RT-Mix to the ready-to-use SYBR® Green RT-PCR master mix and start the reaction.
Shipping and delivery
QuantiNova LNA PCR Assays are shipped at ambient temperatures. In-stock assays are delivered within 1–5 days. During the early access period, longer delivery times should be expected.
What you need to get started with the QuantiNova LNA PCR system
Reverse transcription: QuantiNova Reverse Transcription Kit
qPCR mastermix: QuantiNova SYBR® Green PCR Kit
One-Step qRT-PCR (optional): QuantiNova SYBR® Green RT-PCR Kit
Assays or panels:
- QuantiNova LNA PCR Assays
- QuantiNova LNA PCR Reference Assays
- QuantiNova LNA PCR Custom Assays
- QuantiNova LNA PCR Focus Panels
- QuantiNova LNA PCR lncRNA Focus Panels
- QuantiNova LNA PCR Custom Panels
- QuantiNova LNA PCR Flexible Panels
Applications
QuantiNova LNA PCR Assays are highly suited for applications including:
- mRNA and lncRNA expression analysis, profiling and quantification
- Validation of RNA-seq gene expression data
- Gene expression profiling
- Signal and pathway analysis
- Confirming gene expression knockdown by LNA GapmeRs or siRNAs
- Biomarker development, including screening, identification and validation of disease-associated biomarkers
- Monitoring phenotypic changes related to gene expression
Supporting data and figures
Demonstrated lot-to-lot reproducibility.