Products

QIAsymphony DSP Circulating DNA Kit (192)
Cat. No. / ID: 937556

QIAsymphony DSP Circulating DNA Kit (96)
Cat. No. / ID: 937555

QIAsymphony DSP Circulating DNA Maxi Kit (192)
Cat. No. / ID: 937566
Features
- For in vitro diagnostic use
- Lot-to-lot traceability
- Produced under GMP manufacturing conditions
Product Details
The QIAsymphony DSP Circulating DNA Kits provide reagent cartridges and reagents for fully automated and simultaneous purification of human circulating cell-free (ccf) DNA from human plasma and urine using the QIAsymphony SP instrument. Protocols are available from 1–10 mL sample volumes.
| QIAsymphony DSP Circulating DNA Kit | QIAsymphony DSP Circulating DNA Kit | QIAsymphony DSP Circulating DNA Maxi Kit | |
| Samples | 96 | 192 | 192 |
| Cat. no. | 937555 | 937556 | 937566 |
| Number of reactions | 96 (2 mL, 4 mL, 6 mL, 8 mL and 10 mL sample volume) 192 (1 mL sample volume) |
192 (2 mL and 4 mL sample volume) 384 (1 mL sample volume) |
192 (6 mL, 8 mL and 10 mL sample volume) |
| Reagent cartridge | 2 | 2 | 2 |
| QIAGEN Proteinase K | 3 x 10 mL* | 6 x 10 mL | 13 x 10 mL |
* Additional Proteinase K (cat. no. 19134) bottles should be ordered for 6 mL, 8 mL and 10 mL sample volume to process 96 samples in total.
Principle
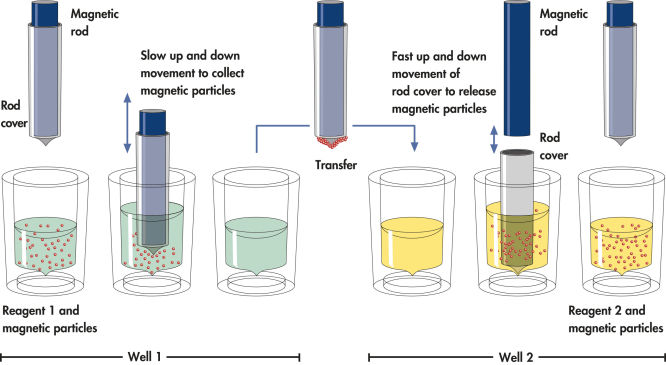
QIAsymphony technology combines the speed and efficiency of anion exchange-based nucleic acid purification with the convenient handling of magnetic particles (see figure “ Schematic diagram of the QIAsymphony SP principle.”).
The QIAsymphony SP processes a sample containing magnetic particles as follows: A magnetic rod protected by a rod cover enters a well containing the sample and attracts the magnetic particles. The magnetic rod cover is positioned above another well and the magnetic particles are released. These steps are repeated several times during sample processing The QIAsymphony SP uses a magnetic head containing an array of 24 magnetic rods, and can therefore process up to 24 samples simultaneously.
See figures
Procedure
The QIAsymphony SP makes automated sample preparation easy and convenient. Samples, reagents and consumables, and eluates are separated in different drawers. Simply load samples, proteinase K, reagents provided in special cartridges, and preracked consumables in the appropriate drawer before a run. Start the protocol and remove purified DNA from the “Eluate” drawer after processing. Refer to the user manuals supplied with your instrument for operating instructions.
The purification procedure is designed to ensure safe and reproducible handling of potentially infectious samples, and comprises 3 steps: bind, wash, and elute (see figure “ QIAsymphony DSP Circulating DNA procedure.”). The user can choose between different sample input volumes.
To process 6–10 mL plasma or urine with the QIAsymphony DSP Circulating DNA Kit (96) (cat. no. 937555), additional Proteinase K (10 mL) (cat. no. 19134). must be purchased.
See figures
Applications
ccfDNA purified using the QIAsymphony DSP Circulating DNA Kit and the QIAsymphony SP is ready for use in a wide range of downstream applications, including NGS, digital, multiplex and quantitative real-time PCR.
Supporting data and figures
Efficient processing of magnetic particles.