✓ Traitement automatique des commandes en ligne 24 h/24 7 j/7
✓ Assistance technique et produits pertinente et professionnelle
✓ Commande (ou réapprovisionnement) rapide et fiable
QIAquick PCR Purification Kit (50)
Cat. No. / ID: 28104
✓ Traitement automatique des commandes en ligne 24 h/24 7 j/7
✓ Assistance technique et produits pertinente et professionnelle
✓ Commande (ou réapprovisionnement) rapide et fiable
Caractéristiques
- Jusqu’à 95 % de récupération d’ADN prêt à l’emploi
- Nettoyage d’ADN jusqu’à 10 kb en trois étapes simples
- Colorant de chargement gélifié pour simplifier l’analyse d’échantillon
- Kit deux-en-un pour l’extraction sur gel et le nettoyage par PCR
- Colonnes de centrifugation disponibles séparément
Détails produit
Le QIAquick PCR Purification Kit contient des colonnes de centrifugation, tampons et tubes de prélèvement pour la purification sur membrane de silice de produits de PCR > 100 pb. L’ADN jusqu’à 10 kb est purifié par une procédure simple et rapide de liaison-lavage-élution, avec un volume d’élution de 30 à 50 μl. Un indicateur de pH facultatif simplifie la détermination du pH optimal pour la liaison de l’ADN à la colonne de centrifugation. La procédure peut être entièrement automatisée sur le système QIAcube Connect.
Pour des résultats optimaux, il est recommandé d’utiliser ce produit avec le QIAvac 24 Plus.
Les protocoles standard QIAquick PCR Purification peuvent aussi être exécutés avec le système TRACKMAN Connected associé aux pipettes PIPETMAN M Connected de Gilson. Le système TRACKMAN Connected guide les chercheurs parmi les protocoles QIAquick PCR Purification tout en ajustant automatiquement les paramètres des pipettes PIPETMAN M Connected activées en Bluetooth. Chaque étape de l’exécution du protocole est enregistrée pour accélérer la création d’un rapport d’exécution complet. Télécharger plus d’informations.
Performances
Voir les illustrations
Principe
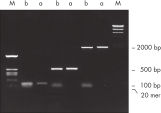
Les QIAquick Kits contiennent une membrane de silice permettant la liaison de l’ADN dans un tampon de forte salinité et l’élution avec un tampon de faible salinité ou de l’eau. La procédure de purification permet d’éliminer les amorces, les nucléotides, les enzymes, l’huile minérale, les sels, l’agarose, le bromure d’éthidium et d’autres impuretés des échantillons d’ADN (voir l’illustration « Élimination complète des amorces après PCR »). La technologie à membrane de silice permet d’écarter les problèmes liés aux résines et aux bouillies non homogènes. Des tampons de liaison spéciaux sont optimisés pour des applications spécifiques et favorisent l’adsorption sélective de molécules d’ADN de tailles bien précises.
Colorant de chargement gélifié
Afin de faciliter un traitement et une analyse plus rapides et pratiques des échantillons, un colorant de chargement gélifié est fourni. Le colorant de chargement GelPilot contient trois colorants de suivi (xylène cyanol, bleu de bromophénol et orange G) pour faciliter l’optimisation du temps d’exécution du gel d’agarose et empêcher les petits fragments d’ADN de migrer trop loin (voir l’illustration « Colorant de chargement GelPilot »).
Voir les illustrations
Procédure
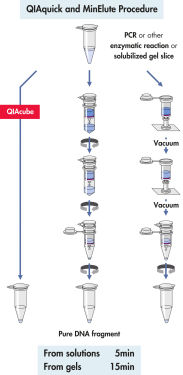
Le système QIAquick utilise une procédure simple de liaison-lavage-élution (voir le schéma « Procédure QIAquick et MinElute »). Le tampon de liaison est ajouté directement à l’échantillon de PCR ou à la réaction enzymatique, puis le mélange est appliqué à la colonne de centrifugation QIAquick. Le tampon de liaison contient un indicateur de pH, cela simplifie la détermination du pH optimal pour la liaison de l’ADN (voir l’illustration « Colorant indicateur de pH »). Les acides nucléiques se fixent à la membrane de silice dans les conditions de forte salinité du tampon. Les impuretés sont éliminées et l’ADN pur est élué dans un petit volume de tampon de faible salinité ou d’eau, prêt à être utilisé dans toutes les autres applications.
Manipulation
Les QIAquick Spin Columns offrent deux possibilités de manipulation pratiques. Les colonnes de centrifugation peuvent être insérées dans une microcentrifugeuse de paillasse classique ou dans n’importe quelle rampe à vide dotée de connecteurs Luer, de type QIAvac 24 Plus avec QIAvac Luer Adapters. Le QIAquick PCR Purification Kit, tout comme d’autres kits QIAGEN avec colonnes de centrifugation, peut être entièrement automatisé sur le QIAcube, cela permet d’accroître la productivité et de standardiser les résultats (voir les illustrations « Possibilités de manipulation des colonnes de centrifugation A, B, C et D » et « QIAcube Connect »).
Voir les illustrations
 Colorant indicateur de pH.
Colorant indicateur de pH.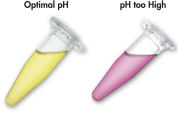 Possibilités de manipulation des colonnes de centrifugation – A.
Possibilités de manipulation des colonnes de centrifugation – A. Possibilités de manipulation des colonnes de centrifugation – B.
Possibilités de manipulation des colonnes de centrifugation – B. Possibilités de manipulation des colonnes de centrifugation – C.
Possibilités de manipulation des colonnes de centrifugation – C.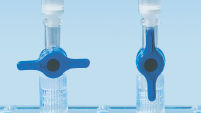 Possibilités de manipulation des colonnes de centrifugation – D.
Possibilités de manipulation des colonnes de centrifugation – D. QIAcube Connect.
QIAcube Connect.
Applications
Les fragments d’ADN purifiés avec le système QIAquick peuvent s’utiliser directement dans toutes les applications, y compris le séquençage, l’analyse de puce à ADN, la ligature et la transformation, la digestion par enzymes de restriction, le marquage, la micro-injection, la PCR et la transcription in vitro.
Données et illustrations utiles
Procédure QIAquick et MinElute.
Specifications
| Features | Specifications |
|---|---|
| Binding capacity | 10 µg |
| Technology | Technologie à base de silice |
| Sample type: applications | ADNsb ou ADNdb à partir de PCR et autres réactions enzymatiques |
| Fragments removed | < 40 mères |
| Elution volume | > 30 µl |
| Fragment size | 100 pb – 10 kb |
| Processing | Manuel |
| Format | Tube |
| Type(s) of DNA recovered | ADNsb et ADNdb |