miRCURY LNA miRNA PCR Assay
Cat. No. / ID: 339306
Features
- Full miRBase coverage enables miRNA profiling from any organism
- Unmatched sensitivity allows miRNA quantification from just 1 pg total RNA
- High specificity discriminates closely related miRNAs and mature miRNA from precursors
- Fast and simple two-step dPCR protocol takes less than 3 hours
- Absolute quantification of miRNA expression changes with dPCR using the QIAcuity instrument and the QIAcuity EG PCR Kit
Product Details
miRCURY LNA miRNA PCR Assays are individual miRNA PCR primer sets that enable extremely sensitive and specific miRNA quantification. Both forward and reverse PCR amplification primers are miRNA-specific and are optimized with LNA technology. The assays are provided in tube format and contain sufficient primers for 166 reactions in a Nanoplate 8.5k (12 μl per reaction) or 50 reactions in a Nanoplate 26k (40 μl per reaction). The upstream cDNA synthesis step is performed with the miRCURY LNA RT Kit, while the digital PCR amplification takes place on the QIAcuity instrument with the QIAcuity EG PCR Kit.
Need a quote for your research project or would you like to discuss your project with our specialist team? Just contact us!
Performance
Optimized digital PCR analysis with the QIAcuity EG PCR Kits and the QIAcuity instrument
miRCURY LNA miRNA PCR Assays were developed to provide high-specificity target amplification. This can be quantified by two PCR methods: qPCR with standard real-time thermocyclers and digital PCR using the QIAcuity instrument and the QIAcuity EG PCR Kit. After cDNA synthesis using the miRCURY LNA RT Kit, digital PCR provides highly precise target quantification, detecting even the smallest expression changes at the lowest concentrations. A subset of key miRNAs have been experimentally validated on the QIAcuity instrument.
LNA enhances PCR performance
miRCURY LNA miRNA PCR Assays have been developed using stringent design criteria and lab-validated algorithms. The rigorous assay selection process ensures that each primer set delivers the highest specificity and efficiency for the most reliable and accurate results. Tm-normalization and LNA enhancement give the primers a higher binding affinity than standard DNA primers, dramatically increasing assay sensitivity and specificity.
Compared with other miRNA PCR systems that use either stem-loop or standard DNA primers, the LNA-enhanced primers offer significantly increased sensitivity, especially for AT-rich miRNAs
Principle
A unique system for miRNA profiling
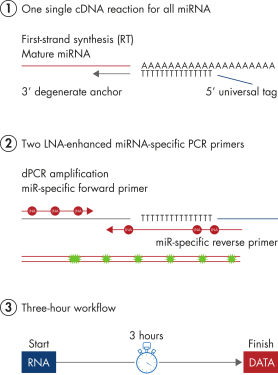
miRCURY LNA miRNA PCR Assays offer the best combination of performance and ease-of-use on the microRNA PCR market by combining universal RT with LNA PCR amplification (see figure Schematic outline of the miRCURY LNA miRNA digital PCR system). Universal RT makes it possible to use one first-strand cDNA synthesis reaction as the template for multiple miRNA real-time PCR assays. This saves precious samples, reduces technical variation and saves time in the laboratory. Plus, both the forward and reverse PCR amplification primers are miRNA specific and optimized with LNA. This provides 1) exceptional sensitivity and extremely low background, enabling accurate quantitation of very low miRNA levels and 2) highly specific assays that allow discrimination between closely related miRNA sequences.
The principle of the dPCR reaction in the QIAcuity Nanoplates is described here.
Coverage
Over 20,000 assays are available covering all organisms in miRBase 20. Over 1,200 assays are fully wet-lab validated for sensitivity, specificity, efficiency and background on both synthetic as well as different biological samples. The remaining assays are in silico-validated using a comprehensive design algorithm that ensures high-quality, species-specific, LNA-enhanced assays with optimal sensitivity and specificity within each organism. This means that several different assays may target the same sequence. Ultimately, the assay for each species is selected based on the genetic background of the organism. If you are working with novel miRNAs, such as from an NGS experiment, custom-designed LNA miRNA primer sets for any miRNA are also available.
Reference gene assays for normalization
Five validated reference gene primer sets are available for dPCR, enabling high-quality data normalization and generation of reliable data from many different sample types (see table below). These control primer sets target endogenous, small non-coding RNAs that are constitutively and relatively stably expressed across different human tissues (see figure Expression levels of reference genes in different human tissues). The control reference genes offer the possibility to accurately and reliably normalize across a range of miRNA expression levels. For more information refer to the miRCURY PCR Controls page.
List of available reference gene assays
| Product name | Target organism | Alternative names | NCBI reference |
| Control primer set, SNORD38B (hsa)* | hsa | U38B; RNU38B | NR_001457 |
| Control primer set, SNORD44 (hsa) | hsa | U44; RNU44 | NR_002750 |
| Control primer set, SNORD48 (hsa) | hsa | U48; RNU48 | NR_002745 |
| Control primer set, SNORD49A (hsa)* | hsa | U49; U49A; RNU49 | NR_002744 |
| Control primer set, SNORA66 (hsa) | hsa | U66; RNU66 | NR_002444 |
| Control primer set, 5S rRNA (hsa) | hsa, mmu | V00589 | |
| Control primer set, U6 snRNA (hsa, mmu)* | hsa, mmu, rno | U6 | X59362 |
| Control primer set, RNU5G snRNA (mmu, hsa)* | mmu, hsa, rno | Rnu5a; U5a; Rnu5g | NR_002852 |
| Control primer set, RNU1A1 (mmu, hsa)* | mmu, hsa, rno | Rnu1a-1; Rnu1a1 | NR_004411 |
| Control primer set, SNORD65 (mmu) | mmu | U65; RNU65 | NR_028541 |
| Control primer set, SNORD68 (mmu) | mmu | U68; RNU68 | NR_028128 |
| Control primer set, SNORD110 (mmu) | mmu | U110; RNU110 | NR_028547 |
* Validated for use in digital PCR.
See figures
Procedure
A simple and rapid plate-based workflow
Partitioning, thermocycling and imaging are all integrated into a single fully automated instrument, delivering results in under two hours. Nanoplate formats are amenable to front-end automation for sample preparation, further reducing hands-on time.
Just like in qPCR experiments, sample preparation includes the transfer of diluted samples and the addition of master mix and primers to a 96- or 24-well nanoplate. The system integrates partitioning, thermocycling and imaging into a single fully automated instrument that takes users from the sample to result in under 3 hours. One can perform remote analysis on the Suite Software, which provides the concentration in copies per microliter of your target sequences as well as in-built quality control options.
Applications
miRCURY LNA miRNA PCR Assays, in combination with the QIAcuity Digital PCR System, enable digital PCR analysis for mature miRNA quantification.
Supporting data and figures
Schematic outline of the miRCURY LNA miRNA digital PCR system.