✓ Procesamiento automático sin interrupción de pedidos en línea
✓ Servicio técnico y para productos experto y profesional
✓ Realización y repetición de pedidos rápidas y fiables
QIAquick Nucleotide Removal Kit (250)
Cat. No. / ID: 28306
✓ Procesamiento automático sin interrupción de pedidos en línea
✓ Servicio técnico y para productos experto y profesional
✓ Realización y repetición de pedidos rápidas y fiables
Características
- Hasta 95 % de recuperación de ADN listo para usarse
- Procedimiento rápido y práctico
- Limpieza de ADN de hasta 10 kb en tres sencillos pasos
- Colorante de carga de gel para el análisis práctico de muestras
Detalles del producto
El QIAquick Nucleotide Removal Kit incluye columnas de centrifugación, tampones y tubos de recogida para la purificación basada en membrana de gel de sílice de nucleótidos y ADN. Se eliminan los nucleótidos no incorporados, las sales y otros contaminantes y se purifican los oligonucleótidos (>17 nt) y los fragmentos de ADN que oscilan entre 40 bp y 10 kb mediante un procedimiento sencillo y rápido de unión, lavado y elución y un volumen de elución de 30–200 µl. El procedimiento se automatiza por completo en el instrumento QIAcube Connect.
Rendimiento
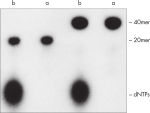
El procedimiento de QIAquick Nucleotide Removal elimina nucleótidos, enzimas, sales y otras impurezas de las muestras de ADN (consulte la figura « Eliminación completa de nucleótidos de los oligonucleótidos marcados»). Con el uso de una microcentrifugadora, se purifica ADN de 17 mer–10 kb. En volúmenes de muestra inferiores a 50 µl, también se puede utilizar el kit DyeEx Spin.
Ver figuras
Principio
Los kits QIAquick contienen un ensamblaje de membrana de sílice para la unión del ADN en tampón de alta salinidad y elución con tampón de baja salinidad o agua. El procedimiento de purificación elimina cebadores, nucleótidos, enzimas, aceites minerales, sales, agarosa, bromuro de etidio y otras impurezas de las muestras de ADN (consulte la figura « Eliminación completa de nucleótidos de los oligonucleótidos marcados»). La tecnología de membrana de sílice elimina los problemas e inconvenientes relacionados con las resinas sueltas y los residuos pastosos. Los tampones de unión especializados están optimizados para aplicaciones específicas y promover la adsorción selectiva de moléculas de ADN dentro de rangos de tamaño específicos.
Colorante de carga de gel
Para que el procesamiento y el análisis de las muestras sean más rápidos y prácticos, se proporciona el colorante de carga de gel. El GelPilot Loading Dye contiene tres colorantes de seguimiento (xileno cianol, azul de bromofenol y naranja G) para facilitar la optimización del tiempo de la serie del gel de agarosa y evitar que los fragmentos más pequeños de ADN migren demasiado lejos (consulte la figura « GelPilot Loading Dye»).
Ver figuras
Procedimiento
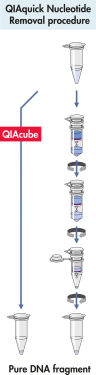
El sistema QIAquick emplea un procedimiento sencillo de unión, lavado y elución (consulte el diagrama de flujo « Procedimiento del QIAquick Nucleotide Removal»). El tampón de unión se añade directamente a la muestra y la mezcla se aplica a la QIAquick Spin Column. Los ácidos nucleicos se adsorben a la membrana de sílice en las condiciones de alta salinidad generadas por el tampón. Se lavan las impurezas y el ADN puro se eluye con un volumen bajo del tampón de baja salinidad suministrado o de agua, listo para usarse en todas las aplicaciones posteriores.
Manejo
Las QIAquick Spin Columns se han diseñado con dos prácticas opciones de manejo. Las columnas de centrifugación se adaptan a una microcentrifugadora de mesa convencional. El QIAquick Nucleotide Removal Kit, junto con otros kits basados en columna de centrifugación de QIAGEN, se puede automatizar por completo en el instrumento QIAcube Connect, lo que permite incrementar la productividad y la normalización de los resultados (consulte las figuras «Opciones de manejo de la columna de centrifugación A» y « QIAcube Connect»).
Ver figuras
Aplicaciones
Los fragmentos de ADN purificados con el sistema QIAquick están listos para su uso directo en diversas aplicaciones, incluyendo la secuenciación, el análisis de micromatrices, la ligación y transformación, la digestión de restricción, el marcado y la microinyección.
Datos y cifras de respaldo
Procedimiento de QIAquick Nucleotide Removal.
Specifications
| Features | Specifications |
|---|---|
| Binding capacity | 10 µg |
| Technology | Tecnología de sílice |
| Recovery: oligonucleotides dsDNA | Recuperación: oligonucleótidos, ADNbc |
| Format | Tubo |
| Elution volume | 30-50 μl |
| Processing | Manual |
| Sample type: applications | ADN, oligonucleótidos: Reacciones de PCR |
| Removal <10mers 17–40mers dye terminator proteins | Eliminación, <10 mers |
| Fragment size | 40 bp–10 kb |