Products

artus M. tuberculosis RG PCR Kit (24) CE
Cat. No. / ID: 4555263

artus M. tuberculosis RG PCR Kit (96) CE
Cat. No. / ID: 4555265
Features
- Compliance with EU IVD Directive 98/79/EC
- High reliability using the internal control
- Highly sensitive detection of as few as 0.23 copies/µl (RG kit)
- Accurate quantitation using the 4 standards supplied
Product Details
Performance
To ensure highest sensitivity, artus M. tuberculosis PCR Kits have been optimized to detect low numbers of mycobacterial DNA. The analytical sensitivity of the artus M. tuberculosis RG PCR Kit is 0.23 copies/µl in the PCR on the Rotor-Gene Q/6000 instrument.
| Kit | artus M. tuberculosis RG PCR Kit |
|---|---|
| Validated sample types | Human sputum, bronchoalveolar lavage (BAL), bronchial secretion, cerebral spinal fluid (CSF), stomach fluid, or peritoneal punction |
| Analytical sensitivity | 0.23 copies/µl in the PCR |
| Specificity | All members of the M. tuberculosis complex (M. tuberculosis, M. africanum, M. bovis, M. bovis BCG, M. microti, and M. pinnipedii) |
Principle
artus M. tuberculosis PCR Kits are based on the amplification and simultaneous detection of a specific region of the mycobacterial genome using real-time PCR. The kits provide high levels of specificity, sensitivity, and reproducibility.
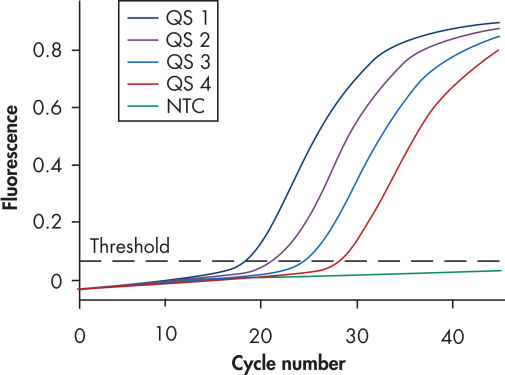
Each artus M. tuberculosis PCR Kit provides 4 M. tuberculosis quantitation standards (see figure " Reliable quantitation of M. tuberculosis complex pathogen load"). Use of the standards enables accurate quantitation of pathogen load. In addition, the kits contain a second heterologous amplification system to identify possible PCR inhibition. This is detected as an internal control (IC) in a different fluorescence channel from the analytical PCR. The detection limit of the analytical M. tuberculosis PCR is not reduced.
| Kit | artus M. tuberculosis RG PCR Kit |
|---|---|
| Validated sample types | Human sputum, bronchoalveolar lavage (BAL), bronchial secretion, cerebral spinal fluid (CSF), stomach fluid, or peritoneal punction |
| Amplicon | 159 bp region of the mycobacterial genome |
See figures
Procedure
artus M. tuberculosis PCR Kits provide all necessary reagents optimized for reliable detection and quantitation of DNA of all members of the M. tuberculosis complex (M. tuberculosis, M. africanum, M. bovis, M. bovis BCG, M. microti, and M. pinnipedii). Simply add template DNA to the ready-to-use PCR master mix and Mg solution, and start the reaction on the appropriate real-time cycler using the optimized cycling program described in the kit handbook.
For DNA purification, the QIAamp DNA Mini Kit is to be used to ensure legally validated performance.
Applications
Supporting data and figures
Reliable quantitation of the M. tuberculosis complex pathogen load.
Specifications
| Features | Specifications |
|---|---|
| Quantitative/qualitative | Quantitative |
| What detected | Mycobacterium tuberculosis DNA (M. tuberculosis, M. africanum, M. bovis...) |
| Recommended sample prep | QIAamp DNA Mini Kit |
| Thermal cycler | ABI PRISM 7000, 7700, 7900HT SDS, LightCycler, Rotor-Gene 3000 |
| RUO/CE/ASR | CE-IVD |
| Sample type | Sputum, bronchoalveolar lavage (BAL), bronchial secretion, cerebrospinal fluid (CSF), stomach fluid, peritoneal punction |