✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
therascreen NRAS Pyro Kit (24)
Cat. No. / ID: 971530
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Compliance with EU IVD Directive 98/79/EC
- Comprehensive results in real time
- Accurate quantification of mutations in the NRAS gene
- Easy interpretation of complex sequence information
Product Details
Performance
Linearity
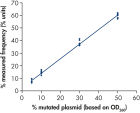
Linearity was determined using mixtures of plasmids carrying the wild-type or mutant sequence for the mutations GGT->GAT in codons 12 and 13 and the mutation CAA->CGA in codon 61 (see figure " Linearity of GGT->GAT mutation"). The plasmids were mixed in proportions to give four levels of mutation (5, 10, 30, and 50%). Each mixture was analyzed with three different lots of the therascreen NRAS Pyro Kit in three Pyrosequencing runs with three replicates each.
The results were linear within an allowable nonlinearity of 5 % units in the tested range of 5 to 50% mutation level. Similar results were obtained for the mutations GGT->GAT in codon 13 and CAA->CGA in codon 61.
Precision
The precision data allows the determination of the total variability of the assays and was obtained at three different levels by analysis of the above mentioned plasmid mixtures with three replicates each.
Repeatability (intra-assay and inter-batch variability) was calculated based on the data for determination of linearity (three runs on the same day using varying lots of the therascreen NRAS Pyro Kit). Intermediate precision (intra laboratory variability) was determined in three runs within one laboratory on three different days with varying operators, PyroMark Q24 instruments, and lots of the therascreen NRAS Pyro Kit. Reproducibility (inter-laboratory variability) was calculated from two runs each in an internal and external laboratory and using varying lots of the therascreen NRAS Pyro Kit.
The repeatability, intermediate precision, and reproducibility for the mutation GGT>GAT in codon 12 was 1.2–1.9, 1.0–2.0, and 1.3–3.1 % units, respectively, in the measured range of 5–50% mutation level. Similar results were obtained for the mutations GGT>GAT in codon 13 and CAA>CGA in codon 61.
| % mutated plasmid | Repeatability (Mean, SD) | Intermediate precision (Mean, SD) | Reproducibility (Mean, SD) |
|---|---|---|---|
| 5 | 7.5, 1.2 | 7.3, 1.0 | 6.7, 1.3 |
| 10 | 14.6, 1.3 | 13.5, 1.1 | 13.7, 1.3 |
| 30 | 37.8, 1.9 | 37.9, 1.5 | 36.1, 2.9 |
| 50 | 59.8, 1.7 | 60.4, 2.0 | 57.5, 3.1 |
See figures
Principle
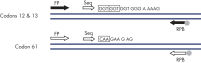
The therascreen NRAS Pyro Kit is used for quantitative measurements of mutations in codons 12, 13, and 61 of the human NRAS gene in real time using Pyrosequencing technology on the PyroMark Q24 System. The NRAS gene encodes a GTPase neuroblastoma RAS viral (v-ras) oncogene homolog. Mutations in NRAS are found in approximately 13–25% of all malignant melanomas, occurring frequently in codons 12, 13, and 61. These mutations results in the constitutive activation of NRAS signaling pathways.
The following mutations are detected:
- Codon 12/13: G12A, G12C, G12D, G12R, G12S, G12V, G13A, G13C, G13D, G13R, G13S, G13V
- Codon 61: Q61E, Q61H, Q61K, Q61L, Q61P, Q61R
Procedure
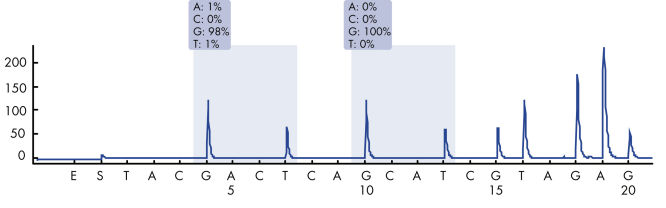
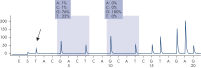
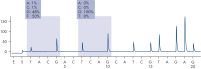
After PCR using primers targeting codons 12/13 and codon 61, the amplicons are immobilized on Streptavidin Sepharose High Performance beads. Single-stranded DNA is prepared, and the corresponding sequencing primers anneal to the DNA. The samples are then analyzed on the PyroMark Q24 System using a run setup file and a run file. The "Sequence to Analyze" can be then adjusted for detection of rare mutations after the run (see figures " Pyrogram trace of a GGT to AGT mutation in base 1 of codon 12" and " Pyrogram trace after reanalysis of the sample in the previous figure").
See figures
Applications
The therascreen NRAS Pyro Kit is used for quantitative measurements of mutations in codons 12, 13, and 61 of the human NRAS gene. The kit is intended to be used as an aid to identify cancer patients more likely to benefit from cancer therapies that act specifically on the NRAS gene or where NRAS is a crucial factor within the pathway.
The following mutations are detected:
- Codon 12: G12A, G12C, G12D, G12R, G12S, G12V
- Codon 13: G13A, G13C, G13D, G13R, G13S, G13V
- Codon 61: Q61E, Q61H, Q61K, Q61L, Q61P, Q61R
Supporting data and figures
Pyrogram trace of a normal genotype in codon 12-13.