✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 206243
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Reliable microsatellite analysis by multiplex PCR
- Microsatellite assay development without optimization
- Successful and specific coamplification of all fragments
- Optimized protocol for fast and reliable results
- Easy instructions for use with various downstream analysis platforms
Product Details
Performance
See figures
Principle
The Type-it Microsatellite PCR Kit provides fast and reliable genotyping analysis of humans, animals, plants, and bacteria using microsatellites, STR, or VNTR markers, without the need for lengthy optimization procedures. The Type-it Microsatellite PCR Kit is based on proprietary QIAGEN multiplex technology and includes highly specific HotStarTaq Plus DNA Polymerase, a chemically modified hot-start enzyme, and a specially developed multiplex buffer system including Factor MP. Together, these components ensure high yields and enable reliable multiplex amplification of all microsatellite loci of interest. The kit includes dedicated protocols for the amplification of microsatellites or STRs, as well as step-by-step advice on template amounts, cycle numbers, and PCR conditions and instrument details for different downstream analysis platforms such as agarose gels, capillary sequencers, the Agilent Bioanalyzer, and the QIAxcel Advanced System.
Type-it Microsatellite PCR Buffer
Unique Type-it Microsatellite PCR Buffer facilitates the amplification of multiple PCR products. In contrast to conventional PCR reagents, the Type-it Microsatellite PCR Buffer contains a specially developed, balanced combination of salts and additives to ensure comparable efficiencies for annealing and extension of all primers in the reaction. Owing to a uniquely balanced combination of KCl and (NH4)2SO4, the PCR buffer provides stringent primer-annealing conditions over a wider range of temperatures and Mg2+ concentrations than conventional PCR buffers. The need to optimize PCR by varying the annealing temperature or the Mg2+ concentration is dramatically reduced, or often not required. Commonly employed optimization procedures for multiplex PCR are virtually eliminated. The buffer also contains the synthetic Factor MP (see figure " Stable and efficient annealing"), which allows efficient primer annealing and extension, irrespective of primer sequence. Factor MP increases the local concentration of primers at the DNA template and stabilizes specifically bound primers.
Q-Solution
The Type-it Microsatellite PCR Kit includes Q-Solution. This innovative PCR additive facilitates amplification of difficult templates by modifying the melting behavior of DNA. Use of this unique reagent will often enable or improve suboptimal PCR. Unlike DMSO and other PCR additives, Q-Solution is used at a defined working concentration with any primer-template system and is not toxic.
See figures
Procedure
Dedicated, application-specific protocols, optimized for analysis of microsatellites on high-resolution capillary sequencers, are included with the Type-it Microsatellite PCR Kit to ensure reliable results for routine analysis, as well as for the establishment of new assays. Reactions can be set up at room temperature, ensuring greater convenience and ease of use.
Fast and simple way to reliable genotyping results
The Type-it Microsatellite PCR Kit enables fast and easy multiplex assay development for accurate and reproducible genotyping results. Whatever the application — microsatellite, STR, SSR, or VNTR analysis — just add the template and primers, and start the thermal-cycler program according to the optimized protocol. The reaction mixture contains all of the reagents required for microsatellite-specific multiplex PCR and enables accurate results without the need for lengthy optimization procedures compared to current methods (see figure " Successful multiplex PCR-based genotyping analysis").
Optimized for use with capillary sequencers
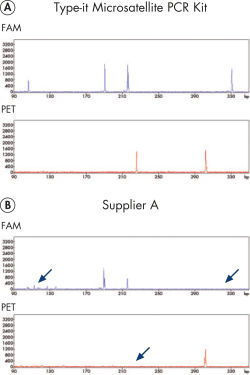
Microsatellite analysis on high-resolution detection systems such as capillary sequencers is often associated with the problem of uneven product yield and huge intensity differences of fluorescent signals. The Type-it Microsatellite PCR Kit overcomes this limitation and ensures high product yields for all amplicons in a multiplex experiment. The kit includes optimized protocols for use with fluorescent primers and outperforms enzyme solutions from other suppliers (see figure " Reliable 13-plex STR analysis without the need for optimization").
See figures
Applications
The Type-it Microsatellite PCR Kit is used to analyze microsatellites, STRs (short tandem repeats), SSRs (simple sequence repeats), and VNTRs (variable number of tandem repeats) and can be used in diverse research fields such as:
- Species or individual identification
- Analysis of relationship
- Microsatellite instability
- Population genetics
Supporting data and figures
Reliable 13-plex STR analysis without the need for optimization.
Specifications
| Features | Specifications |
|---|---|
| Applications | Multiplex PCR, Microsatellites, STRs, VNTRs, SSRs |
| Reaction type | PCR amplification |
| Product use | Functionally validated and developed for reliable Microsatellite Analysis |
| Mastermix | Yes |
| Real-time or endpoint | Endpoint |
| Sample/target type | Genomic DNA |
| Single or multiplex | Multiplex |
| With/without hotstart | With |




