Taq PCR Master Mix Kit
For convenient PCR setup using a premixed solution
For convenient PCR setup using a premixed solution
Cat. No. / ID: 201445
Concentration: 5 units/µl
Recombinant enzyme: Yes
Substrate analogs: dNTP, ddNTP, dUTP, biotin-11-dUTP, DIG-11-dUTP, fluorescent-dNTP/ddNTP
Extension rate: 2–4 kb/min at 72°C
Half-life: 10 min at 97°C; 60 min at 94°C
Amplification efficiency: ≥105 fold
5'–>3' exonuclease activity: Yes
Extra A addition: Yes
3'–>5' exonuclease activity: No
Contaminating nucleases: No
Contaminating RNases: No
Contaminating proteases: No
Self-priming activity: No
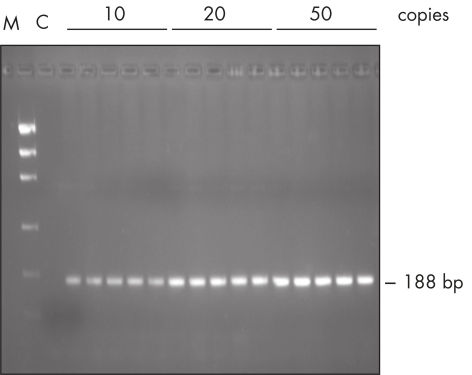
The Taq PCR Master Mix Kit includes QIAGEN's Taq DNA Polymerase in a premixed format. This ready-to-use solution also includes the QIAGEN PCR Buffer, MgCl2, and ultrapure dNTPs at optimized concentrations. Only primers and template DNA need to be added to set up PCR. Due to the convenient master mix format, pipetting errors are minimized, ensuring highly reproducible PCR results (see figure " Reproducible PCR"). Taq PCR Master Mix can be stored at 2–8°C for up to 2 months , allowing even faster PCR setup by eliminating thawing time.
TaqDNA Polymerase DNA Polymerase is a high-quality recombinant enzyme that is suitable for general and specialized PCR applications (see figures " Tolerance of different primer Tm values" and " Specific amplification of long PCR products").
The innovative QIAGEN PCR Buffer has been developed to save time and effort by reducing the need for PCR optimization. QIAGEN PCR Buffer contains both KCl and (NH4)2SO4 (see figure " Increased specificity of primer annealing"). This unique buffer facilitates the amplification of specific PCR products. During the annealing step of every PCR cycle, the buffer allows a high ratio of specific-to-nonspecific primer binding. Owing to a uniquely balanced combination of KCl and (NH4)2SO4, the PCR buffer provides stringent primer-annealing conditions over a wider range of annealing temperatures and Mg2+ concentrations than conventional PCR buffers. Optimization of PCR by varying the annealing temperature or the Mg2+ concentration is dramatically reduced and often not required (see figures " Wide annealing temperature window" and " Tolerance to variable magnesium concentration").
The Taq PCR Master Mix Kit is used for standard and specialized applications, including:
| Features | Specifications |
|---|---|
| Applications | PCR, RT-PCR, DNA fingerprinting |
| dNTP's included | Yes (in Master Mix) |
| Mastermix | Yes |
| Reaction type | PCR amplification |
| Enzyme activity | 5' -> 3' exonuclease activity |
| Real-time or endpoint | Endpoint |
| Sample/target type | Genomic DNA and cDNA |
| Single or multiplex | Single |
| With/without hotstart | Without hotstart |