✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
QIAamp RNA Blood Mini Kit (50)
Cat. No. / ID: 52304
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Rapid purification of high-quality, ready-to-use RNA
- No organic extraction or alcohol precipitation
- Consistent, high yields
- Removal of contaminants and inhibitors
Product Details
The QIAamp RNA Blood Mini Kit provides silica-membrane-based purification of cellular RNA from up to 1.5 ml of fresh, whole human blood stabilized with any common anticoagulant, such as citrate, heparin, or EDTA. After homogenization using the QIAshredder spin column, a fast spin-column procedure simplifies RNA purification. Purification can be fully automated on the QIAcube Connect.
Performance
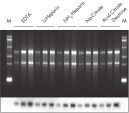
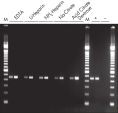
The QIAamp procedure completely removes RNases, contaminants, and enzyme inhibitors, yielding high-quality RNA suitable for any downstream application (see figures " High-quality RNA for northern analysis" and " Reliable RT-PCR analysis").
The QIAamp RNA Blood Mini Kit provides the highest-quality RNA with minimum copurification of DNA. However, as with any RNA purification method, some DNA contamination can be expected. For certain RNA applications that are sensitive to very small amounts of DNA, it may be necessary to remove any remaining DNA. In these cases, the QIAGEN RNase-Free DNase Set provides convenient on-column DNase treatment of RNA samples during QIAamp RNA procedures.
See figures
Principle
The QIAamp RNA Blood Mini Kit provides purification of cellular RNA using silica-membrane technology. No phenol–chloroform extraction is required. RNA binds specifically to the QIAamp silica-gel membrane while contaminants pass through. PCR inhibitors, such as divalent cations and proteins, are completely removed in two efficient wash steps, leaving pure RNA to be eluted in either water or a buffer provided with the kit.
QIAamp technology yields total cellular RNA from fresh whole blood and other sample sources that is ready to use in RT-PCR and blotting procedures. QIAamp sample preparation technology is fully licensed.
Procedure
See figures
Applications
QIAamp RNA Blood Mini Kits provide a fast, easy method for the preparation of total cellular RNA from up to 1.5 ml of human whole blood. Contaminants and enzyme inhibitors, such as hemoglobin and heparin, are completely removed, leaving purified RNA ready for use in downstream applications, such as:
- RT-PCR
- Real-time RT-PCR
- cDNA synthesis
- Northern, dot and slot blotting
- RNase/S1 nuclease protection
- Differential display
- Poly A+ RNA selection
- Primer extension
In addition, QIAamp RNA Blood Mini Kits can be used to purify total RNA from tissues and cultured cells and to separate RNA from proteins, salt and other reaction components after enzymatic reactions, such as DNase digestions, proteinase digestions, RNA ligation and labeling reactions.
Supporting data and figures
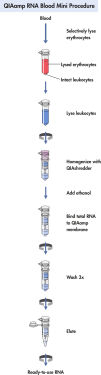
QIAamp RNA Blood Mini procedure.
Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, real-time PCR, microarray |
| Elution volume | 30–100 µl |
| Main sample type | Whole blood, tissue |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein | Cellular RNA |
| Sample amount | 50–1.5 ml |
| Format | Spin columns |
| Processing | Manual (centrifugation) |
| Time per run or per prep | <1 hour |
| Technology | Silica technology |
| Yield | 1–5 µg |