✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
EpiTect Whole Bisulfitome Kit (25)
Cat. No. / ID: 59203
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- High yield of bisulfite converted DNA for multiple PCRs
- Representative amplification from bisulfite converted DNA
- Convenient one-step reaction setup
Product Details
The EpiTect Whole Bisulfitome Kit provides DNA polymerase, buffers, and reagents for amplification of the entire bisulfite converted genome — the bisulfitome — using multiple displacement amplification technology (MDA). The EpiTect Whole Bisulfitome Kit has been specially developed for the amplification of DNA after conversion with EpiTect Bisulfite Kits. It uses proven REPLI-g technology and has been adapted to the specific requirements of bisulfite converted DNA (smaller DNA fragment size and altered nucleotide composition due to bisulfite conversion), while maintaining the converted sequence representation. Typical DNA yields from the EpiTect Whole Bisulfitome Kit are 1–3 μg per reaction, sufficient for up to 300 PCR analyses from just a single EpiTect bisulfite reaction.
Performance
See figures
Principle
Methylation analysis of genomic DNA sequences can be limited by the small amount of sample available. The EpiTect Whole Bisulfitome Kit provides highly uniform amplification across the entire bisulfite converted genome, with negligible sequence bias (see figure " EpiTYPER MALDI-TOF analysis of WBA DNA"). The method is based on isothermal multiple displacement amplification (MDA) technology. A uniquely processive DNA polymerase with a 3'→5' exonuclease proofreading activity maintains high fidelity during the replication process.
The EpiTect Whole Bisulfitome Kit has been specially developed using proven REPLI-g technology and has been adapted to the special requirements of bisulfite converted DNA (smaller DNA fragment size and changed nucleotide composition due to bisulfite conversion), while maintaining the converted sequence representation. Best results are obtained when used in combination with EpiTect Plus Bisulfite Kits, which contain a uniquely formulated DNA Protect system. This enables effective DNA denaturation, which is necessary for complete cytosine conversion, and prevents the excessive DNA fragmentation usually associated with high temperature and low pH conditions of the bisulfite conversion reaction. Preventing DNA fragmentation enables subsequent amplification of large DNA fragments. Typically, DNA yields of 1–3 μg are generated per reaction using the EpiTect Whole Bisulfitome Kit.
See figures
Procedure
The convenient one-step reaction setup involves incubating bisulfite converted template DNA with a mixture of reaction buffer and REPLI-g DNA polymerase. After heat inactivation of the polymerase, the amplified DNA is ready for analysis or storage (see figure " WBA procedure").
Accessing epigenetic information is of prime importance for many areas of biological and medical research — particularly oncology, but also stem cell research and developmental biology. However, the analysis of changes in DNA methylation is challenging due to the lack of standardized methods for providing reproducible data particularly from limited sample material. With its newly introduced EpiTect products, QIAGEN makes available standardized, pre-analytical and analytical solutions, from DNA sample collection, stabilization and purification, to bisulfite conversion and real-time or end-point PCR methylation analysis or sequencing (see figure " Standardized workflows in epigenetics").
See figures
Applications
The EpiTect Whole Bisulfitome Kit is the ideal tool to provide sufficient DNA for all downstream methylation analyses, whenever bisulfite converted DNA is limited. The amplification reaction produces enough DNA for 100–300 real-time PCR assays (e.g., using EpiTect MethyLight PCR Kits) or end-point PCRs (e.g., using EpiTect MSP Kits), estimating 10 ng of amplified DNA per reaction.
Supporting data and figures
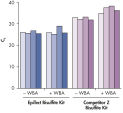
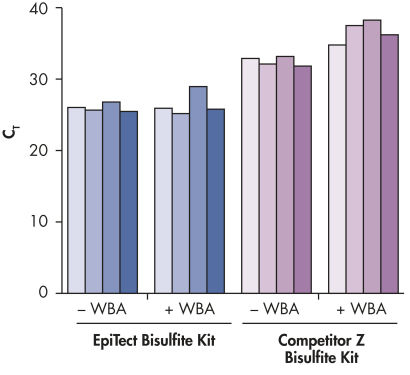
Reliable whole bisulfitome amplification.
Real-time PCR was used to measure the representation of four different loci in 10 ng bisulfite converted DNA, before (–WBA) and after (+WBA) amplification with the EpiTect Whole Bisulfitome Kit. DNA was first bisulfite converted using either the EpiTect Bisulfite Kit or a kit from supplier Z, and was then amplified; qPCR was performed using the QuantiTect SYBR® Green PCR Kit. In the EpiTect converted DNA, the four loci were represented similarly before and after amplification. Higher CT values for the DNA converted with the competitor’s bisulfite kit indicate higher DNA fragmentation prior to WBA. Following amplification, the relative representation of the four loci was increased and different, indicating lower whole bisulfitome amplification and unequal amplification.
Specifications
| Features | Specifications |
|---|---|
| Amplification | Whole bisulfite converted DNA |
| Reaction time | 8h at 28°C |
| Maximum input volume | 10 µl bisulfite converted DNA |
| Starting amount of DNA | > 50 ng bisulfite converted DNA |
| Applications | Methylation specific PCR |
| Reaction volume | 40 µl |
| Starting material | Epitect bisulfite converted DNA |
| Technology | Multiple Displacement Amplification (MDA) |
| Yield | 1-3 µg per reaction |