✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
RNeasy 96 Universal Tissue 8000 Kit (12)
Cat. No. / ID: 967852
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- High yields of total RNA from cells and all tissues
- Fast and convenient sample processing
- Pure, high-quality RNA without phenol contamination
- High-performance RNA for all downstream applications
Product Details
RNeasy 96 BioRobot Kits enable total RNA purification in 96-well format for BioRobot instruments from cultured cell samples and any tissue. Tissues are efficiently lysed and homogenized in QIAzol Lysis Reagent while cells are lyses in Buffer RLT. High-quality RNA is purified from tissues and cells using silica-membrane RNeasy 96 plates. Automated purification is performed using the BioRobot Universal System, which processes up to 96 or 192 samples per run followed by real-time RT-PCR setup. Tissue samples can be conveniently stabilized using Allprotect Tissue Reagent, and efficiently disrupted using the TissueLyser II. Nonfatty tissues can be stabilized using the RNAprotect Tissue Reagent.
Performance
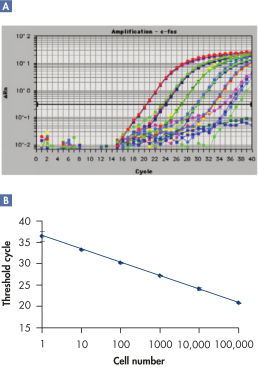
The RNeasy 96 system provides fast and reproducible total RNA purification in 96-well format for BioRobot instruments for high-throughput gene expression profiling. The RNA is suitable for sensitive applications such as quantitative, real-time RT-PCR and microarray analysis (see figure " High-quality RNA for sensitive analysis of a low-copy transcript"). Sample sizes range from 10 to 5 x 105 cells (see figure " RT-PCR of RNA from as few as 100 cells"), and high-quality RNA can be purified from large numbers of samples (see figure " Reproducible yields of high-quality RNA"). Individual variations are low throughout the entire purification process; TaqMan® threshold-cycle values are easily achieved at the end of the process with a coefficient of variation (CV) less than 3% using the QuantiTect Probe RT-PCR Kit (see figure " Repeatability of fully automated RNA purification").
Compared with standard silica-membrane procedures, the RNeasy 96 system provides higher yields of total RNA for all tissue types (see table “Typical total RNA yields using the RNeasy 96 Universal Tissue Kit”).
Typical total RNA yields using the RNeasy 96 Universal Tissue Kit
| Tissue | RNA yield* ((µg per 10 mg of tissue) |
|---|---|
| Kidney | 5–40 |
| Liver | 15–80 |
| Lung | 5–15 |
| Heart | 5–25 |
| Muscle | 5–35 |
| Brain | 5–20 |
| Adipose tissue | 0.5–2.5 |
| Spleen | 15–100 |
| Intestine | 10–60 |
| Skin | 2–5 |
High-quality RNA can be isolated from tissues preserved either by freezing or by stabilization with RNAprotect at room temperature (see figure " High-quality total RNA from rat liver and muscle tissue"). Intact RNA purified from difficult-to-lyse fibrous and fatty tissues is suitable for downstream applications (see figures " High-quality RNA from skin / adipose / liver / intestine tissue" and " Real-time analysis of high-quality RNA from rat brain").
See figures
 RT-PCR of RNA from as few as 100 cells.
RT-PCR of RNA from as few as 100 cells. Reproducible yields of high-quality RNA.
Reproducible yields of high-quality RNA. Repeatability of fully automated RNA purification.
Repeatability of fully automated RNA purification.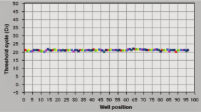 High quality total RNA from rat liver and muscle tissue.
High quality total RNA from rat liver and muscle tissue.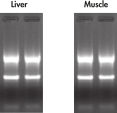 High-quality RNA from skin tissue.
High-quality RNA from skin tissue.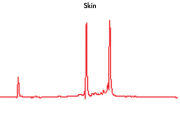 High-quality RNA from adipose tissue.
High-quality RNA from adipose tissue.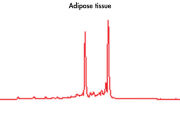 High-quality RNA from liver tissue.
High-quality RNA from liver tissue.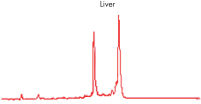 High-quality RNA from intestine tissue.
High-quality RNA from intestine tissue.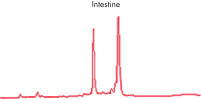 Real-time analysis of high-quality RNA from rat brain.
Real-time analysis of high-quality RNA from rat brain.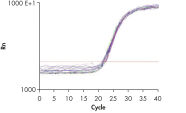
Principle
The RNeasy 96 BioRobot 8000 Kit provides a standardized, fast, reliable method for isolation of high-quality RNA from large numbers of samples (see table “Specifications of the RNeasy 96 BioRobot 8000 Kit”). The simple procedure is key to the speed and efficiency of RNeasy 96 technology. Cells can be grown and directly lysed in 96-well cell-culture dishes. After transfer of the lysates to the wells of the RNeasy 96 plate (see figure " RNeasy 96-well plate"), RNA binds to the silica membrane in each well, and contaminants are washed away. Pure RNA is then eluted in RNase-free water into individual collection tubes suitable for long-term storage and is ready to use for any experiment. Since the RNeasy procedure enriches for RNA species >200 nt, RNA yield does not include 5S rRNA, tRNAs, or other low-molecular-weight RNAs.
Specifications of the RNeasy 96 BioRobot 8000 Kit
| Specification | Details |
|---|---|
| Format | RNeasy 96-well plates with 1.2 ml collection microtubes |
| Sample source | Animal or human cells |
| Sample size | 10–5 x 105 cells |
| Binding capacity | Up to 100 µg RNA per well |
| Yield* from 1 x 105 cells | HeLa: 1.6 µg LMH: 1.3 µg COS-7: 3.1 µg Huh7: 2.0 µg Jurkat: 1.4 µg |
| Elution volume | 45–140 µl |
See figures
Procedure
Fully automated total RNA isolation using the RNeasy 96 BioRobot 8000 Kit is performed on the BioRobot 8000 workstation or BioRobot Universal System. The BioRobot worktable has the capacity to purify RNA from up to 192 samples in a single run. With the one-plate protocol, the system can carry out both RNA purification and reaction setup in a single run – from living cell cultures in 96-well plates to RT-PCR mixtures, ready to use in real-time gene expression analysis. Filter tips prevent cross-contamination, and precision robotic handling provides minimal well-to-well variation and high repeatability. RNA purification can also be performed manually on the QIAvac 96 vacuum manifold and/or the 96-Well-Plate Centrifugation System using the RNeasy 96 Kit.
The RNeasy 96 Universal Tissue 8000 Kit integrates efficient phenol/guanidine-based lysis and silica-membrane purification with the speed of vacuum processing. Following homogenization and phase separation, the BioRobot 8000 workstation or BioRobot Universal System provides walkaway automation of the procedure, for total RNA purification from all types of animal tissue (see flowchart " RNeasy 96 Universal Tissue 8000 procedure").
Tissue is first efficiently lysed using QIAzol Lysis Reagent and the TissueLyser II. This provides rapid and parallel disruption of tissues and inactivation of RNases to ensure purification of intact RNA. After phase separation by centrifugation, the BioRobot workstation removes the aqueous phase and adds ethanol to provide appropriate binding conditions. The workstation then applies the sample to the wells of the RNeasy 96 Universal Tissue plate, where total RNA binds and contaminants are efficiently washed away. High-performance RNA is then eluted in a small volume of water, ready for use in any downstream application.
See figures
Applications
The RNeasy 96 BioRobot 8000 Kit consistently provides the highest-quality RNA for:
- High-throughput RNA applications in gene expression analysis
- Sensitive applications such as quantitative, real-time RT-PCR
- Pharmacological and toxicological research
The combination of QIAzol and RNeasy technologies results in highly pure RNA without phenol carryover. RNA purified using the RNeasy 96 Universal Tissue 8000 Kit is suitable for all downstream applications, including real-time RT-PCR.
Comparison of RNeasy 96 BioRobot Kits
| Features | RNeasy 96 BioRobot 8000 Kit | RNeasy 96 Universal Tissue 8000 Kit |
|---|---|---|
| Applications | PCR, real-time PCR, microarray | PCR, qPCR, real-time PCR |
| Elution volume | 45–140 µl | Custom |
| For automated processing |
BioRobot 8000 Workstation or BioRobot Universal System |
BioRobot 8000 Workstation or BioRobot Universal System |
| Format | 96-well plate | 96-well plate |
| Main sample type | Cultured cells | Tissue samples |
| Processing | Automated | Automated |
|
Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein |
RNA | RNA |
| Sample amount | 5 x 105 | 20–80 mg |
| Technology | Silica technology | Silica technology |
| Yield | 1.3–3.1 µg | Varies |
Supporting data and figures
High-quality RNA for sensitive analysis of a low-copy transcript.