EZ2 AllPrep DNA/RNA FFPE Kit (48)
Cat. No. / ID: 954734
Features
- Patented technology to extract DNA and RNA from the same sample – no need to divide the sample
- Elution of DNA and RNA in separate fractions offers convenient downstream analysis
- Reduces sample consumption
- Optimizes crosslink removal for both DNA and RNA
- No overnight proteinase K incubation
Product Details
The EZ2 AllPrep DNA/RNA FFPE Kit is used with the EZ2 Connect instrument. It automates the extraction of both DNA and RNA, from exactly the same piece of sample in two distinct elution fractions.
The kit technology allows the reversal of crosslinking caused by the sample’s exposure to formalin without overnight proteinase K incubation.
Performance
The EZ2 AllPrep DNA/RNA FFPE Kit is designed for automated purification of DNA and RNA from FFPE samples, using magnetic-bead technology and prefilled cartridges.
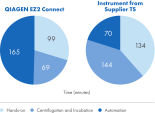
The automated workflow is presented in figure " Automated, simultaneous DNA and RNA purification from FFPE samples." This workflow provides a greater degree of automation (figure
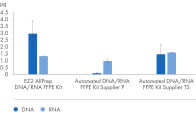
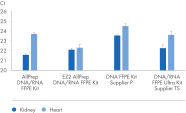
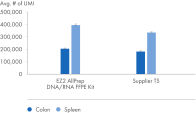
" Degree of automation") and provides superior nucleic acid yields from FFPE samples compared to manual workflows and workflows from other suppliers (figure " DNA and RNA yields from spleen FFPE.", figure " DNA and RNA yields from human spleen.", figure " RNA and DNA yield from rat kidney and heart." and figure " Comparison of manual and automated extraction."). The decrosslinking step improves efficiency and library complexity (figure " Decrosslinking efficiency." and figure
" Library complexity.")
See figures
Principle
DNA and RNA extraction from the same sample is needed to compare genomic and transcriptomic data reliably. Usually, this is done by splitting the sample into two separate reactions/tubes/containers. The extractions are then processed separately. Thus, DNA is extracted from one piece of the sample, while RNA is extracted from the other piece.
Conventional methods have two major disadvantages: First, the amount of nucleic acids that can be extracted from the sample is cut into half. Second, because DNA is extracted from one group of cells, and RNA is extracted from a separate group, there can be differences in the properties of the cell sources.
The patented AllPrep DNA/RNA FFPE procedure incubates FFPE tissue samples in a lysis buffer that releases RNA into the supernatant and leaves DNA in the pellet. The RNA and DNA are then purified in separate tubes but within the same automated run using one cartridge in the EZ2 Connect instrument. This way, DNA and RNA can be isolated from as many as 24 samples, all at the same time.
Crosslink removal
Extended crosslink removal at high temperature reverses more crosslinks, but it also increases nucleic acid fragmentation. RNA is more susceptible to heat-induced fragmentation than DNA – but it is also less affected by crosslinks.
Therefore, the optimal conditions for decrosslinking DNA and RNA are different.
By generating separate lysates for RNA and DNA, the EZ2 AllPrep DNA/RNA FFPE workflow allows optimal crosslink removal conditions for each nucleic acid type.
Procedure
EZ2 AllPrep DNA/RNA FFPE technology solubilizes and incubates a single FFPE sample to release RNA into the supernatant while DNA is left in the pellet. The supernatant is incubated to remove RNA crosslinking. Finally, the tubes that separately contain the supernatant and pellet(s) are transferred to EZ2 Connect.
The EZ2 Connect automates all remaining steps of the workflow. These include an additional lysis and crosslink-removal step for DNA, binding of nucleic acids to magnetic particles, removal of contaminants with multiple washing steps, and elution of DNA and RNA into different tubes (see EZ2 AllPrep DNA/RNA FFPE workflow).
See figures
Applications
DNA and RNA purified with the EZ2 AllPrep DNA/RNA FFPE Kit can be used for real-time PCR, Pyrosequencing and other downstream applications.
Supporting data and figures
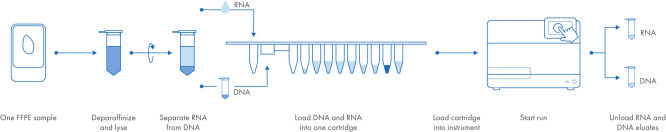
Automated, simultaneous DNA and RNA purification from FFPE samples.
With the EZ2 AllPrep system, DNA and RNA are purified simultaneously from a single sample, using a single workflow, a single cartridge and a single run. The bind, wash and elute steps are fully automated. The workflow is drastically shortened to only a few manual steps. In the EZ2 AllPrep DNA/RNA FFPE Kit workflow, a FFPE sample is placed in a tube, deparaffinized and lysed. The DNA-containing pellet and RNA-containing supernatant are separated by centrifugation. After decrosslinking and DNase treatment of the RNA-containing supernatant, the samples are placed into the same EZ2 cartridge. That cartridge is then loaded into the EZ2 Connect instrument, and the run is started. When the run is finished, the RNA and DNA eluates (in separate tubes) are ready for use in downstream applications.
Specifications
| Features | Specifications |
|---|---|
| Applications | (RT-)PCR, (RT-)qPCR, NGS, genotyping, microarray analysis |
| Elution volume | RNA: 50–60 µl; DNA: 100–120 µl |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein | RNA and DNA |
| Technology | Magnetic particle technology |
| Number of preps per run | 1–24 samples per run |
| Main sample type | FFPE tissue samples |
| Sample amount | max. 4*10 µm sections or 2*20 µm sections |
| Yield | Varies depending on sample type, as well as on storage and fixation conditions |